An organoid-based screen for epigenetic inhibitors that stimulate antigen presentation and potentiate T-cell-mediated cytotoxicity
- PMID: 34725507
- PMCID: PMC8647932
- DOI: 10.1038/s41551-021-00805-x
An organoid-based screen for epigenetic inhibitors that stimulate antigen presentation and potentiate T-cell-mediated cytotoxicity
Erratum in
-
Author Correction: An organoid-based screen for epigenetic inhibitors that stimulate antigen presentation and potentiate T-cell-mediated cytotoxicity.Nat Biomed Eng. 2025 Feb;9(2):281-282. doi: 10.1038/s41551-023-01096-0. Nat Biomed Eng. 2025. PMID: 37648780 No abstract available.
Abstract
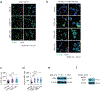
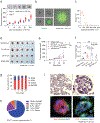
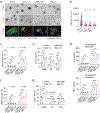
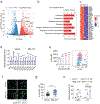
In breast cancer, genetic heterogeneity, the lack of actionable targets and immune evasion all contribute to the limited clinical response rates to immune checkpoint blockade therapy. Here, we report a high-throughput screen based on the functional interaction of mouse- or patient-derived breast tumour organoids and tumour-specific cytotoxic T cells for the identification of epigenetic inhibitors that promote antigen presentation and potentiate T-cell-mediated cytotoxicity. We show that the epigenetic inhibitors GSK-LSD1, CUDC-101 and BML-210, identified by the screen, display antitumour activities in orthotopic mammary tumours in mice, that they upregulate antigen presentation mediated by the major histocompatibility complex class I on breast tumour cells and that treatment with BML-210 substantially sensitized breast tumours to the inhibitor of the checkpoint programmed death-1. Standardized measurements of tumour-cell killing activity facilitated by tumour-organoid-T-cell screens may help with the identification of candidate immunotherapeutics for a range of cancers.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
XL and XZ are submitting a patent for the organoid-based screen method and identified drugs in this study.
Figures











References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

