Metformin induces muscle atrophy by transcriptional regulation of myostatin via HDAC6 and FoxO3a
- PMID: 34725961
- PMCID: PMC8818615
- DOI: 10.1002/jcsm.12833
Metformin induces muscle atrophy by transcriptional regulation of myostatin via HDAC6 and FoxO3a
Abstract
Background: Skeletal muscle atrophy is a severe condition that involves loss of muscle mass and quality. Drug intake can also cause muscle atrophy. Biguanide metformin is the first-line and most widely prescribed anti-diabetic drug for patients with type 2 diabetes. The molecular mechanism of metformin in muscle is unclear.
Methods: Myostatin expression was investigated at the protein and transcript levels after metformin administration. To investigate the pathways associated with myostatin signalling, we used real-time polymerase chain reaction, immunoblotting, luciferase assay, chromatin immunoprecipitation assay, co-immunoprecipitation, immunofluorescence, primary culture, and confocal microscopy. Serum analysis, physical performance, and immunohistochemistry were performed using our in vivo model.
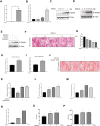
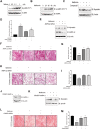
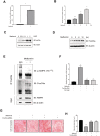
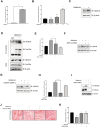
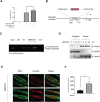
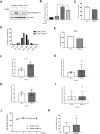
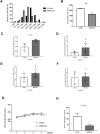
Results: Metformin induced the expression of myostatin, a key molecule that regulates muscle volume and triggers the phosphorylation of AMPK. AMPK alpha2 knockdown in the background of metformin treatment reduced the myostatin expression of C2C12 myotubes (-49.86 ± 12.03%, P < 0.01) and resulted in increased myotube diameter compared with metformin (+46.62 ± 0.88%, P < 0.001). Metformin induced the interaction between AMPK and FoxO3a, a key transcription factor of myostatin. Metformin also altered the histone deacetylase activity in muscle cells (>3.12-fold ± 0.13, P < 0.001). The interaction between HDAC6 and FoxO3a induced after metformin treatment. Confocal microscopy revealed that metformin increased the nuclear localization of FoxO3a (>3.3-fold, P < 0.001). Chromatin immunoprecipitation revealed that metformin induced the binding of FoxO3a to the myostatin promoter. The transcript-level expression of myostatin was higher in the gastrocnemius (GC) muscles of metformin-treated wild-type (WT) (+68.9 ± 10.01%, P < 0.001) and db/db mice (+55.84 ± 6.62%, P < 0.001) than that in the GC of controls (n = 4 per group). Average fibre cross-sectional area data also showed that the metformin-treated C57BL/6J (WT) (-31.74 ± 0.75%, P < 0.001) and C57BLKS/J-db/db (-18.11 ± 0.94%, P < 0.001) mice had decreased fibre size of GC compared to the controls. The serum myoglobin level was significantly decreased in metformin-treated WT mice (-66.6 ± 9.03%, P < 0.01).
Conclusions: Our results demonstrate that metformin treatment impairs muscle function through the regulation of myostatin in skeletal muscle cells via AMPK-FoxO3a-HDAC6 axis. The muscle-wasting effect of metformin is more evident in WT than in db/db mice, indicating that more complicated mechanisms may be involved in metformin-mediated muscular dysfunction.
Keywords: AMPK; FoxO3a; HDAC6; Metformin; Muscle atrophy; Myostatin.
© 2021 The Authors. Journal of Cachexia, Sarcopenia and Muscle published by John Wiley & Sons Ltd on behalf of Society on Sarcopenia, Cachexia and Wasting Disorders.
Conflict of interest statement
All authors declare that they have no conflicts of interest.
Figures
References
-
- Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J 2013;280:4294–4314. - PubMed
-
- Hoogaars WMH, Jaspers RT. Past, present, and future perspective of targeting myostatin and related signaling pathways to counteract muscle atrophy. Adv Exp Med Biol 2018;1088:153–206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

