The crystal structures of the enzyme hydroxymethylbilane synthase, also known as porphobilinogen deaminase
- PMID: 34726177
- PMCID: PMC8561815
- DOI: 10.1107/S2053230X2100964X
The crystal structures of the enzyme hydroxymethylbilane synthase, also known as porphobilinogen deaminase
Abstract
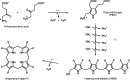
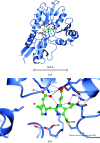
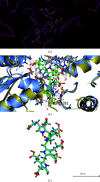
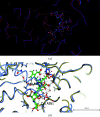
The enzyme hydroxymethylbilane synthase (HMBS; EC 4.3.1.8), also known as porphobilinogen deaminase, catalyses the stepwise addition of four molecules of porphobilinogen to form the linear tetrapyrrole 1-hydroxymethylbilane. Thirty years of crystal structures are surveyed in this topical review. These crystal structures aim at the elucidation of the structural basis of the complex reaction mechanism involving the formation of tetrapyrrole from individual porphobilinogen units. The consistency between the various structures is assessed. This includes an evaluation of the precision of each molecular model and what was not modelled. A survey is also made of the crystallization conditions used in the context of the operational pH of the enzyme. The combination of 3D structural techniques, seeking accuracy, has also been a feature of this research effort. Thus, SAXS, NMR and computational molecular dynamics have also been applied. The general framework is also a considerable chemistry research effort to understand the function of the enzyme and its medical pathologies in acute intermittent porphyria (AIP). Mutational studies and their impact on the catalytic reaction provide insight into the basis of AIP and are also invaluable for guiding the understanding of the crystal structure results. Future directions for research on HMBS are described, including the need to determine the protonation states of key amino-acid residues identified as being catalytically important. The question remains - what is the molecular engine for this complex reaction? Thermal fluctuations are the only suggestion thus far.
Keywords: enzyme–substrate intermediates; hydroxymethylbilane synthase; porphobilinogen deaminase; reaction mechanisms; structure and function.
open access.
Figures


References
-
- Bung, N., Roy, A., Priyakumar, U. D. & Bulusu, G. (2019). Phys. Chem. Chem. Phys. 21, 7932–7940. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

