Membrane nanodomains modulate formin condensation for actin remodeling in Arabidopsis innate immune responses
- PMID: 34726756
- PMCID: PMC8774048
- DOI: 10.1093/plcell/koab261
Membrane nanodomains modulate formin condensation for actin remodeling in Arabidopsis innate immune responses
Abstract
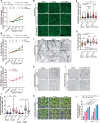
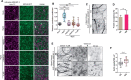
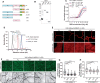
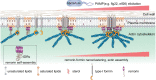
The assembly of macromolecules on the plasma membrane concentrates cell surface biomolecules into nanometer- to micrometer-scale clusters (nano- or microdomains) that help the cell initiate or respond to signals. In plant-microbe interactions, the actin cytoskeleton undergoes rapid remodeling during pathogen-associated molecular pattern-triggered immunity (PTI). The nanoclustering of formin-actin nucleator proteins at the cell surface has been identified as underlying actin nucleation during plant innate immune responses. Here, we show that the condensation of nanodomain constituents and the self-assembly of remorin proteins enables this mechanism of controlling formin condensation and activity during innate immunity in Arabidopsis thaliana. Through intrinsically disordered region-mediated remorin oligomerization and formin interaction, remorin gradually recruits and condenses formins upon PTI activation in lipid bilayers, consequently increasing actin nucleation in a time-dependent manner postinfection. Such nanodomain- and remorin-mediated regulation of plant surface biomolecules is expected to be a general feature of plant innate immune responses that creates spatially separated biochemical compartments and fine tunes membrane physicochemical properties for transduction of immune signals in the host.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Comment in
-
Back to the roots: A focus on plant cell biology.Plant Cell. 2022 Jan 20;34(1):1-3. doi: 10.1093/plcell/koab278. Plant Cell. 2022. PMID: 34755878 Free PMC article. No abstract available.
-
Connecting the dots: Membrane nanodomains mediate clustering of actin-nucleator Type I formins in Arabidopsis immune responses.Plant Cell. 2022 Jan 20;34(1):6-7. doi: 10.1093/plcell/koab277. Plant Cell. 2022. PMID: 35226747 Free PMC article. No abstract available.
References
-
- Abel NB, Buschle CA, Hernandez-Ryes C, Burkart SS, Deroubaix A-F, Mergner J, Gronnier J, Jarsch IK, Folgmann J, Braun KH (2021) A hetero-oligomeric remorin-receptor complex regulates plant development. bioRxiv. DOI: 10.1101/2021.01.28.428596 [Posted on January 29, 2021]
-
- Bariola P, Retelska D, Stasiak A, Kammerer R, Fleming A, Hijri M, Frank S, Farmer E (2004) Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol Biol 55: 579–594 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

