CRISPR-dependent endogenous gene regulation is required for virulence in piscine Streptococcus agalactiae
- PMID: 34727007
- PMCID: PMC8592606
- DOI: 10.1080/22221751.2021.2002127
CRISPR-dependent endogenous gene regulation is required for virulence in piscine Streptococcus agalactiae
Abstract
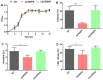
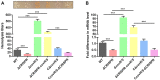
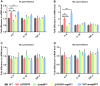
The clustered regularly interspaced palindromic repeats (CRISPR)-Cas (CRISPR-associated) system is a prokaryotic defence against invading mobile genetic elements, such as bacteriophages or exogenous plasmids. Beyond this, this system has been shown to play an important role in controlling the virulence of some bacterial pathogens. Streptococcus agalactiae strain GD201008-001, a causative agent of septicemia and meningitis in tilapia, contains a single type II CRISPR-Cas system with Cas9 as a signature protein. In this study, we found that the deletion of CRISPR significantly reduced adhesion, invasion, cytotoxicity and haemolysis, and caused severely attenuated virulence in the piscine S. agalactiae strain. RNA-Seq identified 236 endogenous genes regulated by CRISPR, with 159 genes upregulated and 77 genes downregulated. The resulting change in gene transcription by CRISPR was much more pronounced than that by cas9 in this bacterium, indicating CRISPR-mediated endogenous gene regulation was mostly independently of cas9. Subsequent studies showed that CovR/S two-component system was transcriptionally upregulated due to CRISPR deletion, which repressed the expression of the cylE gene coding for a cytolytic toxin, and thus decreased the activity of β-haemolysin/cytolysin. However, upregulation of CovR/S was not the contributor to the attenuation phenotype of ΔCRISPR. Further, we demonstrated that CRISPR is capable of repressing the expression of Toll-like receptor 2 (TLR2)-activating lipoprotein Sag0671 and thus dampens the innate immune response. This study revealed that the CRISPR system of S. agalactiae exhibited extraordinary potential capability in the regulation of endogenous transcripts, which contributes to bacterial innate immune evasion and virulence.
Keywords: CRISPR; CovR/S; Streptococcus agalactiae; lipoprotein Sag0671; virulence.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
-
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526(7571):55–61. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
