Profiling the specificity of clonally expanded plasma cells during chronic viral infection by single-cell analysis
- PMID: 34727578
- PMCID: PMC9299196
- DOI: 10.1002/eji.202149331
Profiling the specificity of clonally expanded plasma cells during chronic viral infection by single-cell analysis
Abstract
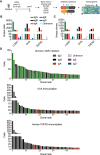
Plasma cells and their secreted antibodies play a central role in the long-term protection against chronic viral infection. However, due to experimental limitations, a comprehensive description of linked genotypic, phenotypic, and antibody repertoire features of plasma cells (gene expression, clonal frequency, virus specificity, and affinity) has been challenging to obtain. To address this, we performed single-cell transcriptome and antibody repertoire sequencing of the murine BM plasma cell population following chronic lymphocytic choriomeningitis virus infection. Our single-cell sequencing approach recovered full-length and paired heavy- and light-chain sequence information for thousands of plasma cells and enabled us to perform recombinant antibody expression and specificity screening. Antibody repertoire analysis revealed that, relative to protein immunization, chronic infection led to increased levels of clonal expansion, class-switching, and somatic variants. Furthermore, antibodies from the highly expanded and class-switched (IgG) plasma cells were found to be specific for multiple viral antigens and a subset of clones exhibited cross-reactivity to nonviral and autoantigens. Integrating single-cell transcriptome data with antibody specificity suggested that plasma cell transcriptional phenotype was correlated to viral antigen specificity. Our findings demonstrate that chronic viral infection can induce and sustain plasma cell clonal expansion, combined with significant somatic hypermutation, and can generate cross-reactive antibodies.
Keywords: LCMV; immune receptor repertoire sequencing; plasma cells; single-cell; viral infection.
© 2021 The Authors. European Journal of Immunology published by Wiley-VCH GmbH.
Conflict of interest statement
Authors declare that there are no conflict of interest.
Figures





References
-
- Greczmiel, U. , Kräutler, N. J. , Pedrioli, A. , Bartsch, I. , Agnellini, P. , Bedenikovic, G. , Harker, J. et al., Sustained T follicular helper cell response is essential for control of chronic viral infection. Sci. Immunol. 2017. 2: eaam8686. - PubMed
-
- Battegay, M. , Moskophidis, D. , Waldner, H. , Bründler, M. A. , Fung‐Leung, W. P. , Mak, T. W. , Hengartner, H. et al., Impairment and delay of neutralizing antiviral antibody responses by virus‐specific cytotoxic T cells. J. Immunol. 1993. 151: 5408–5415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

