Extracellular LGALS3BP regulates neural progenitor position and relates to human cortical complexity
- PMID: 34728600
- PMCID: PMC8564519
- DOI: 10.1038/s41467-021-26447-w
Extracellular LGALS3BP regulates neural progenitor position and relates to human cortical complexity
Abstract
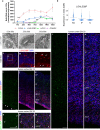
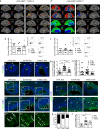
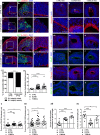
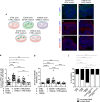
Basal progenitors (BPs), including intermediate progenitors and basal radial glia, are generated from apical radial glia and are enriched in gyrencephalic species like humans, contributing to neuronal expansion. Shortly after generation, BPs delaminate towards the subventricular zone, where they further proliferate before differentiation. Gene expression alterations involved in BP delamination and function in humans are poorly understood. Here, we study the role of LGALS3BP, so far known as a cancer biomarker, which is a secreted protein enriched in human neural progenitors (NPCs). We show that individuals with LGALS3BP de novo variants exhibit altered local gyrification, sulcal depth, surface area and thickness in their cortex. Additionally, using cerebral organoids, human fetal tissues and mice, we show that LGALS3BP regulates the position of NPCs. Single-cell RNA-sequencing and proteomics reveal that LGALS3BP-mediated mechanisms involve the extracellular matrix in NPCs' anchoring and migration within the human brain. We propose that its temporal expression influences NPCs' delamination, corticogenesis and gyrification extrinsically.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

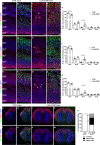

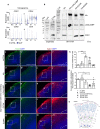
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

