A gene toolbox for monitoring autophagy transcription
- PMID: 34728604
- PMCID: PMC8563709
- DOI: 10.1038/s41419-021-04121-9
A gene toolbox for monitoring autophagy transcription
Abstract
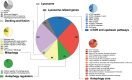
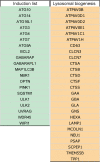
Autophagy is a highly dynamic and multi-step process, regulated by many functional protein units. Here, we have built up a comprehensive and up-to-date annotated gene list for the autophagy pathway, by combining previously published gene lists and the most recent publications in the field. We identified 604 genes and created main categories: MTOR and upstream pathways, autophagy core, autophagy transcription factors, mitophagy, docking and fusion, lysosome and lysosome-related genes. We then classified such genes in sub-groups, based on their functions or on their sub-cellular localization. Moreover, we have curated two shorter sub-lists to predict the extent of autophagy activation and/or lysosomal biogenesis; we next validated the "induction list" by Real-time PCR in cell lines during fasting or MTOR inhibition, identifying ATG14, ATG7, NBR1, ULK1, ULK2, and WDR45, as minimal transcriptional targets. We also demonstrated that our list of autophagy genes can be particularly useful during an effective RNA-sequencing analysis. Thus, we propose our lists as a useful toolbox for performing an informative and functionally-prognostic gene scan of autophagy steps.
© 2021. The Author(s).
Conflict of interest statement
Andrea Ballabio is a Co-Founder of CASMA Therapeutics.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

