High-throughput imaging of ATG9A distribution as a diagnostic functional assay for adaptor protein complex 4-associated hereditary spastic paraplegia
- PMID: 34729478
- PMCID: PMC8557665
- DOI: 10.1093/braincomms/fcab221
High-throughput imaging of ATG9A distribution as a diagnostic functional assay for adaptor protein complex 4-associated hereditary spastic paraplegia
Abstract
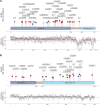
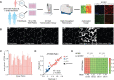
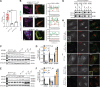
Adaptor protein complex 4-associated hereditary spastic paraplegia is caused by biallelic loss-of-function variants in AP4B1, AP4M1, AP4E1 or AP4S1, which constitute the four subunits of this obligate complex. While the diagnosis of adaptor protein complex 4-associated hereditary spastic paraplegia relies on molecular testing, the interpretation of novel missense variants remains challenging. Here, we address this diagnostic gap by using patient-derived fibroblasts to establish a functional assay that measures the subcellular localization of ATG9A, a transmembrane protein that is sorted by adaptor protein complex 4. Using automated high-throughput microscopy, we determine the ratio of the ATG9A fluorescence in the trans-Golgi-network versus cytoplasm and ascertain that this metric meets standards for screening assays (Z'-factor robust >0.3, strictly standardized mean difference >3). The 'ATG9A ratio' is increased in fibroblasts of 18 well-characterized adaptor protein complex 4-associated hereditary spastic paraplegia patients [mean: 1.54 ± 0.13 versus 1.21 ± 0.05 (standard deviation) in controls] and receiver-operating characteristic analysis demonstrates robust diagnostic power (area under the curve: 0.85, 95% confidence interval: 0.849-0.852). Using fibroblasts from two individuals with atypical clinical features and novel biallelic missense variants of unknown significance in AP4B1, we show that our assay can reliably detect adaptor protein complex 4 function. Our findings establish the 'ATG9A ratio' as a diagnostic marker of adaptor protein complex 4-associated hereditary spastic paraplegia.
Keywords: adaptor protein complex 4; biomarker; functional assay; hereditary spastic paraplegia; high-throughput imaging.
© The Author(s) (2021). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures




References
-
- Shribman S, Reid E, Crosby AH, Houlden H, Warner TT.. Hereditary spastic paraplegia: From diagnosis to emerging therapeutic approaches. Lancet Neurol. 2019;18(12):1136–1146. - PubMed
-
- Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. - PMC - PubMed
-
- Ebrahimi-Fakhari D, Behne R, Davies AK, Hirst J, et al.AP-4-associated hereditary spastic paraplegia. In: Adam MP, Ardinger HH, Pagon RA, eds. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 2018, 1993–2021. PMID: 30543385. - PubMed
-
- Sanger A, Hirst J, Davies AK, Robinson MS.. Adaptor protein complexes and disease at a glance. J Cell Sci. 2019;132(20):jcs222992. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
