Characterization of the Interaction Domains between the Phosphoprotein and the Nucleoprotein of Human Metapneumovirus
- PMID: 34730389
- PMCID: PMC8791268
- DOI: 10.1128/JVI.00909-21
Characterization of the Interaction Domains between the Phosphoprotein and the Nucleoprotein of Human Metapneumovirus
Abstract
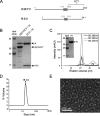
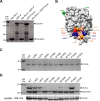
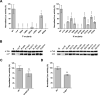
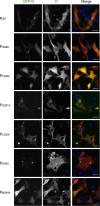
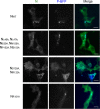
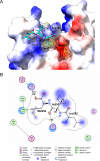
Human metapneumovirus (HMPV) causes severe respiratory diseases in young children. The HMPV RNA genome is encapsidated by the viral nucleoprotein (N), forming an RNA-N complex (NNuc), which serves as the template for genome replication and mRNA transcription by the RNA-dependent RNA polymerase (RdRp). The RdRp is formed by the association of the large polymerase subunit (L), which has RNA polymerase, capping, and methyltransferase activities, and the tetrameric phosphoprotein (P). P plays a central role in the RdRp complex by binding to NNuc and L, allowing the attachment of the L polymerase to the NNuc template. During infection these proteins concentrate in cytoplasmic inclusion bodies (IBs) where viral RNA synthesis occurs. By analogy to the closely related pneumovirus respiratory syncytial virus (RSV), it is likely that the formation of IBs depends on the interaction between HMPV P and NNuc, which has not been demonstrated yet. Here, we finely characterized the binding P-NNuc interaction domains by using recombinant proteins, combined with a functional assay for the polymerase complex activity, and the study of the recruitment of these proteins to IBs by immunofluorescence. We show that the last 6 C-terminal residues of HMPV P are necessary and sufficient for binding to NNuc and that P binds to the N-terminal domain of N (NNTD), and we identified conserved N residues critical for the interaction. Our results allowed us to propose a structural model for the HMPV P-NNuc interaction. IMPORTANCE Human metapneumovirus (HMPV) is a leading cause of severe respiratory infections in children but also affects human populations of all ages worldwide. Currently, no vaccine or efficient antiviral treatments are available for this pneumovirus. A better understanding of the molecular mechanisms involved in viral replication could help the design or discovery of specific antiviral compounds. In this work, we have investigated the interaction between two major viral proteins involved in HMPV RNA synthesis, the N and P proteins. We finely characterized their domains of interaction and identified a pocket on the surface of the N protein, a potential target of choice for the design of compounds interfering with N-P complexes and inhibiting viral replication.
Keywords: HMPV; nucleoprotein; phosphoprotein; protein-protein interaction; structural modeling.
Figures



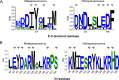
References
-
- Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand F-X, Briese T, Bukreyev A, Calisher CH, Chandran K, Chéng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astúa J, Formenty P, Fouchier RAM, Fù Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li C-X, Lin X-D, Liú L, Longdon B, Marton S, Maisner A, Mühlberger E, Netesov SV, Nowotny N, et al.. 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. 10.1007/s00705-016-2880-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

