Targeting glioblastoma signaling and metabolism with a re-purposed brain-penetrant drug
- PMID: 34731610
- PMCID: PMC8856626
- DOI: 10.1016/j.celrep.2021.109957
Targeting glioblastoma signaling and metabolism with a re-purposed brain-penetrant drug
Abstract
The highly lethal brain cancer glioblastoma (GBM) poses a daunting challenge because the blood-brain barrier renders potentially druggable amplified or mutated oncoproteins relatively inaccessible. Here, we identify sphingomyelin phosphodiesterase 1 (SMPD1), an enzyme that regulates the conversion of sphingomyelin to ceramide, as an actionable drug target in GBM. We show that the highly brain-penetrant antidepressant fluoxetine potently inhibits SMPD1 activity, killing GBMs, through inhibition of epidermal growth factor receptor (EGFR) signaling and via activation of lysosomal stress. Combining fluoxetine with temozolomide, a standard of care for GBM, causes massive increases in GBM cell death and complete tumor regression in mice. Incorporation of real-world evidence from electronic medical records from insurance databases reveals significantly increased survival in GBM patients treated with fluoxetine, which was not seen in patients treated with other selective serotonin reuptake inhibitor (SSRI) antidepressants. These results nominate the repurposing of fluoxetine as a potentially safe and promising therapy for patients with GBM and suggest prospective randomized clinical trials.
Keywords: EGFR signaling; Membrane lipids; SMPD1; combination therapy; electronic medical records; fluoxetine; glioblastoma; real-world evidence; sphingolipid metabolism.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.S.M. is co-founder of Boundless Bio, Inc. He has equity in the company, chairs the scientific advisory board, and serves as a consultant, for which he is compensated. P.S.M. is also consultant for Autobahn Therapeutics, Inc. and Sage Therapeutics. These consulting arrangements started after completion of this paper.
Figures

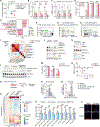




References
-
- Andersen PK, and Gill RD (1982). Cox regression-model for counting-processes: a large sample study. Ann. Stat. 10, 1100–1120.
-
- Bi J, Chowdhry S, Wu S, Zhang W, Masui K, and Mischel PS (2020). Altered cellular metabolism in gliomas - an emerging landscape of actionable co-dependency targets. Nat. Rev. Cancer 20, 57–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 NS073366/NS/NINDS NIH HHS/United States
- R01 CA238249/CA/NCI NIH HHS/United States
- R00 NS082381/NS/NINDS NIH HHS/United States
- P50 HD103557/HD/NICHD NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- R01 NS073831/NS/NINDS NIH HHS/United States
- K99 NS082381/NS/NINDS NIH HHS/United States
- K02 AG068617/AG/NIA NIH HHS/United States
- P50 CA211015/CA/NCI NIH HHS/United States
- P30 HD004612/HD/NICHD NIH HHS/United States
- R01 NS080939/NS/NINDS NIH HHS/United States
- R56 NS080939/NS/NINDS NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

