Fibroblast growth factor receptor fusions in cancer: opportunities and challenges
- PMID: 34732230
- PMCID: PMC8564965
- DOI: 10.1186/s13046-021-02156-6
Fibroblast growth factor receptor fusions in cancer: opportunities and challenges
Abstract
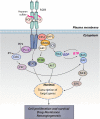
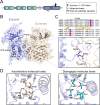
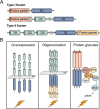
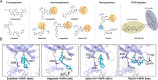
Fibroblast growth factors (FGFs) and their receptors (FGFRs) play critical roles in many biological processes and developmental functions. Chromosomal translocation of FGFRs result in the formation of chimeric FGFR fusion proteins, which often cause aberrant signaling leading to the development and progression of human cancer. Due to the high recurrence rate and carcinogenicity, oncogenic FGFR gene fusions have been identified as promising therapeutic targets. Erdafitinib and pemigatinib, two FGFR selective inhibitors targeting FGFR fusions, have been approved by the U.S. Food and Drug Administration (FDA) to treat patients with urothelial cancer and cholangiocarcinoma, respectively. Futibatinib, a third-generation FGFR inhibitor, is under phase III clinical trials in patients with FGFR gene rearrangements. Herein, we review the current understanding of the FGF/FGFRs system and the oncogenic effect of FGFR fusions, summarize promising inhibitors under clinical development for patients with FGFR fusions, and highlight the challenges in this field.
Keywords: Cancer; Chromosomal translocation; Fibroblast growth factor receptors; Fusion proteins; Inhibitors.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

