Metabolic and Metabo-Clinical Signatures of Type 2 Diabetes, Obesity, Retinopathy, and Dyslipidemia
- PMID: 34732537
- PMCID: PMC8914294
- DOI: 10.2337/db21-0490
Metabolic and Metabo-Clinical Signatures of Type 2 Diabetes, Obesity, Retinopathy, and Dyslipidemia
Abstract
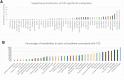
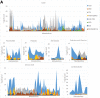
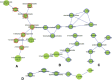
Macro- and microvascular complications of type 2 diabetes (T2D), obesity, and dyslipidemia share common metabolic pathways. In this study, using a total of 1,300 metabolites from 996 Qatari adults (57% with T2D) and 1,159 metabolites from an independent cohort of 2,618 individuals from the Qatar BioBank (11% with T2D), we identified 373 metabolites associated with T2D, obesity, retinopathy, dyslipidemia, and lipoprotein levels, 161 of which were novel. Novel metabolites included phospholipids, sphingolipids, lysolipids, fatty acids, dipeptides, and metabolites of the urea cycle and xanthine, steroid, and glutathione metabolism. The identified metabolites enrich pathways of oxidative stress, lipotoxicity, glucotoxicity, and proteolysis. Second, we identified 15 patterns we defined as "metabo-clinical signatures." These are clusters of patients with T2D who group together based on metabolite levels and reveal the same clustering in two or more clinical variables (obesity, LDL, HDL, triglycerides, and retinopathy). These signatures revealed metabolic pathways associated with different clinical patterns and identified patients with extreme (very high/low) clinical variables associated with extreme metabolite levels in specific pathways. Among our novel findings are the role of N-acetylmethionine in retinopathy in conjunction with dyslipidemia and the possible roles of N-acetylvaline and pyroglutamine in association with high cholesterol levels and kidney function.
© 2022 by the American Diabetes Association.
Figures






References
-
- Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 2007;167:1068–1074 - PubMed
-
- Krauss RM. Lipids and lipoproteins in patients with type 2 diabetes. Diabetes Care 2004;27:1496–1504 - PubMed
-
- Ng TW, Khan AA, Meikle PJ. Investigating the pathogenesis and risk of type 2 diabetes: clinical applications of metabolomics. Clin Lipidol 2012;7:641–659
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

