Gene-level analysis of rare variants in 379,066 whole exome sequences identifies an association of GIGYF1 loss of function with type 2 diabetes
- PMID: 34732801
- PMCID: PMC8566487
- DOI: 10.1038/s41598-021-99091-5
Gene-level analysis of rare variants in 379,066 whole exome sequences identifies an association of GIGYF1 loss of function with type 2 diabetes
Abstract
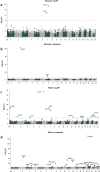
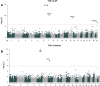
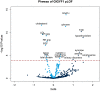
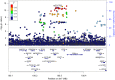
Sequencing of large cohorts offers an unprecedented opportunity to identify rare genetic variants and to find novel contributors to human disease. We used gene-based collapsing tests to identify genes associated with glucose, HbA1c and type 2 diabetes (T2D) diagnosis in 379,066 exome-sequenced participants in the UK Biobank. We identified associations for variants in GCK, HNF1A and PDX1, which are known to be involved in Mendelian forms of diabetes. Notably, we uncovered novel associations for GIGYF1, a gene not previously implicated by human genetics in diabetes. GIGYF1 predicted loss of function (pLOF) variants associated with increased levels of glucose (0.77 mmol/L increase, p = 4.42 × 10-12) and HbA1c (4.33 mmol/mol, p = 1.28 × 10-14) as well as T2D diagnosis (OR = 4.15, p = 6.14 × 10-11). Multiple rare variants contributed to these associations, including singleton variants. GIGYF1 pLOF also associated with decreased cholesterol levels as well as an increased risk of hypothyroidism. The association of GIGYF1 pLOF with T2D diagnosis replicated in an independent cohort from the Geisinger Health System. In addition, a common variant association for glucose and T2D was identified at the GIGYF1 locus. Our results highlight the role of GIGYF1 in regulating insulin signaling and protecting from diabetes.
© 2021. The Author(s).
Conflict of interest statement
A.D, L.W., M.P., A.F.C., L.B., G.H. and P.N. are employees and stockholders of Alnylam Pharmaceuticals. P.A., L.L. and A.B. are employees and stockholders of Regeneron Pharmaceuticals.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

