Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression
- PMID: 34732891
- PMCID: PMC8612926
- DOI: 10.1038/s41586-021-04062-5
Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression
Erratum in
-
Author Correction: Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression.Nature. 2023 Aug;620(7976):E29. doi: 10.1038/s41586-023-06523-5. Nature. 2023. PMID: 37580531 Free PMC article. No abstract available.
Abstract
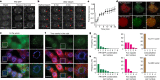
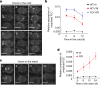
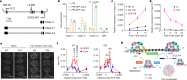
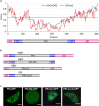
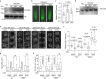
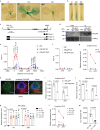
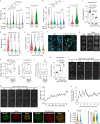
Plants use seasonal temperature cues to time the transition to reproduction. In Arabidopsis thaliana, winter cold epigenetically silences the floral repressor locus FLOWERING LOCUS C (FLC) through POLYCOMB REPRESSIVE COMPLEX 2 (PRC2)1. This vernalization process aligns flowering with spring. A prerequisite for silencing is transcriptional downregulation of FLC, but how this occurs in the fluctuating temperature regimes of autumn is unknown2-4. Transcriptional repression correlates with decreased local levels of histone H3 trimethylation at K36 (H3K36me3) and H3 trimethylation at K4 (H3K4me3)5,6, which are deposited during FRIGIDA (FRI)-dependent activation of FLC7-10. Here we show that cold rapidly promotes the formation of FRI nuclear condensates that do not colocalize with an active FLC locus. This correlates with reduced FRI occupancy at the FLC promoter and FLC repression. Warm temperature spikes reverse this process, buffering FLC shutdown to prevent premature flowering. The accumulation of condensates in the cold is affected by specific co-transcriptional regulators and cold induction of a specific isoform of the antisense RNA COOLAIR5,11. Our work describes the dynamic partitioning of a transcriptional activator conferring plasticity in response to natural temperature fluctuations, thus enabling plants to effectively monitor seasonal progression.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Cold induction of nuclear FRIGIDA condensation in Arabidopsis.Nature. 2023 Jul;619(7969):E27-E32. doi: 10.1038/s41586-023-06189-z. Epub 2023 Jul 12. Nature. 2023. PMID: 37438599 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

