Analysis of the Replication Mechanisms of the Human Papillomavirus Genomes
- PMID: 34733254
- PMCID: PMC8558456
- DOI: 10.3389/fmicb.2021.738125
Analysis of the Replication Mechanisms of the Human Papillomavirus Genomes
Abstract
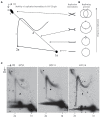
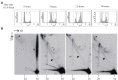
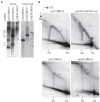
The life-cycle of human papillomaviruses (HPVs) includes three distinct phases of the viral genome replication. First, the viral genome is amplified in the infected cells, and this amplification is often accompanied by the oligomerization of the viral genomes. Second stage includes the replication of viral genomes in concert with the host cell genome. The viral genome is further amplified during the third stage of the viral-life cycle, which takes place only in the differentiated keratinocytes. We have previously shown that the HPV18 genomes utilize at least two distinct replication mechanisms during the initial amplification. One of these mechanisms is a well-described bidirectional replication via theta type of replication intermediates. The nature of another replication mechanism utilized by HPV18 involves most likely recombination-dependent replication. In this paper, we show that the usage of different replication mechanisms is a property shared also by other HPV types, namely HPV11 and HPV5. We further show that the emergence of the recombination dependent replication coincides with the oligomerization of the viral genomes and is dependent on the replicative DNA polymerases. We also show that the oligomeric genomes of HPV18 replicate almost exclusively using recombination dependent mechanism, whereas monomeric HPV31 genomes replicate bi-directionally during the maintenance phase of the viral life-cycle.
Keywords: 2D; HPV; non-theta replication; theta replication; virus replication.
Copyright © 2021 Liblekas, Piirsoo, Laanemets, Tombak, Laaneväli, Ustav, Ustav and Piirsoo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Astell C. R., Chow M. B., Ward D. C. (1985). Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J. Virol. 54, 171–177. doi: 10.1128/jvi.54.1.171-177.1985, PMID: - DOI - PMC - PubMed
-
- Auborn K. J., Little R. D., Platt T. H. K., Vaccariello M. A., Schildkraut C. L. (1994). Replicative intermediates of human papillomavirus type 11 in laryngeal papillomas: site of replication initiation and direction of replication. Proc. Natl. Acad. Sci. U. S. A. 91, 7340–7344. doi: 10.1073/pnas.91.15.7340, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

