HI-NESS: a family of genetically encoded DNA labels based on a bacterial nucleoid-associated protein
- PMID: 34734265
- PMCID: PMC8789088
- DOI: 10.1093/nar/gkab993
HI-NESS: a family of genetically encoded DNA labels based on a bacterial nucleoid-associated protein
Abstract
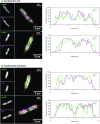
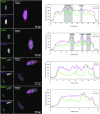
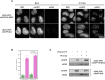
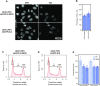
The interplay between three-dimensional chromosome organisation and genomic processes such as replication and transcription necessitates in vivo studies of chromosome dynamics. Fluorescent organic dyes are often used for chromosome labelling in vivo. The mode of binding of these dyes to DNA cause its distortion, elongation, and partial unwinding. The structural changes induce DNA damage and interfere with the binding dynamics of chromatin-associated proteins, consequently perturbing gene expression, genome replication, and cell cycle progression. We have developed a minimally-perturbing, genetically encoded fluorescent DNA label consisting of a (photo-switchable) fluorescent protein fused to the DNA-binding domain of H-NS - a bacterial nucleoid-associated protein. We show that this DNA label, abbreviated as HI-NESS (H-NS-based indicator for nucleic acid stainings), is minimally-perturbing to genomic processes and labels chromosomes in eukaryotic cells in culture, and in zebrafish embryos with preferential binding to AT-rich chromatin.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
The Origin of Chromosomal Replication Is Asymmetrically Positioned on the Mycobacterial Nucleoid, and the Timing of Its Firing Depends on HupB.J Bacteriol. 2018 Apr 24;200(10):e00044-18. doi: 10.1128/JB.00044-18. Print 2018 May 15. J Bacteriol. 2018. PMID: 29531181 Free PMC article.
-
HupB Is a Bacterial Nucleoid-Associated Protein with an Indispensable Eukaryotic-Like Tail.mBio. 2017 Nov 7;8(6):e01272-17. doi: 10.1128/mBio.01272-17. mBio. 2017. PMID: 29114022 Free PMC article.
-
[Molecular mechanisms involved in bacterial chromatin organization].Postepy Biochem. 2019 Oct 1;65(3):202-211. doi: 10.18388/pb.2019_270. Postepy Biochem. 2019. PMID: 31643167 Review. Polish.
-
Structure and function of bacterial H-NS protein.Biochem Soc Trans. 2016 Dec 15;44(6):1561-1569. doi: 10.1042/BST20160190. Biochem Soc Trans. 2016. PMID: 27913665 Review.
-
Building bridges within the bacterial chromosome.Trends Genet. 2015 Mar;31(3):164-73. doi: 10.1016/j.tig.2015.01.003. Epub 2015 Feb 11. Trends Genet. 2015. PMID: 25682183 Review.
Cited by
-
Cell wall damage increases macromolecular crowding effects in the Escherichia coli cytoplasm.iScience. 2023 Mar 9;26(4):106367. doi: 10.1016/j.isci.2023.106367. eCollection 2023 Apr 21. iScience. 2023. PMID: 37009215 Free PMC article.
-
FRET-FISH probes chromatin compaction at individual genomic loci in single cells.Nat Commun. 2022 Nov 5;13(1):6680. doi: 10.1038/s41467-022-34183-y. Nat Commun. 2022. PMID: 36335096 Free PMC article.
References
-
- Dame R.T., Rashid F.Z.M., Grainger D.C.. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet. 2020; 21:227–242. - PubMed
-
- Dame R.T., Tark-Dame M.. Bacterial chromatin: converging views at different scales. Curr. Opin. Cell Biol. 2016; 40:60–65. - PubMed
-
- Banerjee D., Pal S.K.. Dynamics in the DNA recognition by DAPI: exploration of the various binding modes. J. Phys. Chem. B. 2008; 112:1016–1021. - PubMed
-
- Trotta E., D’Ambrosio E., Ravagnan G., Paci M. Simultaneous and different binding mechanisms of 4′,6-diamidino-2-phenylindole to DNA hexamer (d(CGATCG))2: A 1H NMR study. J. Biol. Chem. 1996; 271:27608–27614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

