Plant PhysioSpace: a robust tool to compare stress response across plant species
- PMID: 34734276
- PMCID: PMC8566309
- DOI: 10.1093/plphys/kiab325
Plant PhysioSpace: a robust tool to compare stress response across plant species
Abstract
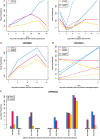
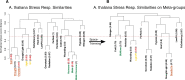
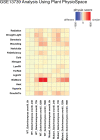
Generalization of transcriptomics results can be achieved by comparison across experiments. This generalization is based on integration of interrelated transcriptomics studies into a compendium. Such a focus on the bigger picture enables both characterizations of the fate of an organism and distinction between generic and specific responses. Numerous methods for analyzing transcriptomics datasets exist. Yet, most of these methods focus on gene-wise dimension reduction to obtain marker genes and gene sets for, for example, pathway analysis. Relying only on isolated biological modules might result in missing important confounders and relevant contexts. We developed a method called Plant PhysioSpace, which enables researchers to compute experimental conditions across species and platforms without a priori reducing the reference information to specific gene sets. Plant PhysioSpace extracts physiologically relevant signatures from a reference dataset (i.e. a collection of public datasets) by integrating and transforming heterogeneous reference gene expression data into a set of physiology-specific patterns. New experimental data can be mapped to these patterns, resulting in similarity scores between the acquired data and the extracted compendium. Because of its robustness against platform bias and noise, Plant PhysioSpace can function as an inter-species or cross-platform similarity measure. We have demonstrated its success in translating stress responses between different species and platforms, including single-cell technologies. We have also implemented two R packages, one software and one data package, and a Shiny web application to facilitate access to our method and precomputed models.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Bergman NH (2007) Comparative Genomics. Humana Press, Springer Science & Business Media, Totowa, NJ https://www.ncbi.nlm.nih.gov/books/NBK1732/ - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

