Modeling Metastatic Colonization in a Decellularized Organ Scaffold-Based Perfusion Bioreactor
- PMID: 34734500
- PMCID: PMC11469127
- DOI: 10.1002/adhm.202100684
Modeling Metastatic Colonization in a Decellularized Organ Scaffold-Based Perfusion Bioreactor
Abstract
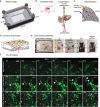
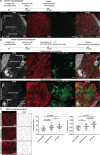
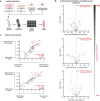
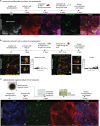
Metastatic cancer spread is responsible for most cancer-related deaths. To colonize a new organ, invading cells adapt to, and remodel, the local extracellular matrix (ECM), a network of proteins and proteoglycans underpinning all tissues, and a critical regulator of homeostasis and disease. However, there is a major lack in tools to study cancer cell behavior within native 3D ECM. Here, an in-house designed bioreactor, where mouse organ ECM scaffolds are perfused and populated with cells that are challenged to colonize it, is presented. Using a specialized bioreactor chamber, it is possible to monitor cell behavior microscopically (e.g., proliferation, migration) within the organ scaffold. Cancer cells in this system recapitulate cell signaling observed in vivo and remodel complex native ECM. Moreover, the bioreactors are compatible with co-culturing cell types of different genetic origin comprising the normal and tumor microenvironment. This degree of experimental flexibility in an organ-specific and 3D context, opens new possibilities to study cell-cell and cell-ECM interplay and to model diseases in a controllable organ-specific system ex vivo.
Keywords: cancer metastasis; experimental methods; extracellular matrix; specialized bioreactors.
© 2021 The Authors. Advanced Healthcare Materials published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Discher D. E., Janmey P., Wang Y. L., Science 2005, 310, 1139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

