Genome mining methods to discover bioactive natural products
- PMID: 34734626
- PMCID: PMC8597713
- DOI: 10.1039/d1np00032b
Genome mining methods to discover bioactive natural products
Abstract
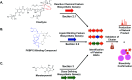
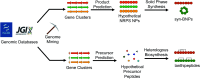
Covering: 2016 to 2021With genetic information available for hundreds of thousands of organisms in publicly accessible databases, scientists have an unprecedented opportunity to meticulously survey the diversity and inner workings of life. The natural product research community has harnessed this breadth of sequence information to mine microbes, plants, and animals for biosynthetic enzymes capable of producing bioactive compounds. Several orthogonal genome mining strategies have been developed in recent years to target specific chemical features or biological properties of bioactive molecules using biosynthetic, resistance, or transporter proteins. These "biosynthetic hooks" allow researchers to query for biosynthetic gene clusters with a high probability of encoding previously undiscovered, bioactive compounds. This review highlights recent case studies that feature orthogonal approaches that exploit genomic information to specifically discover bioactive natural products and their gene clusters.
Conflict of interest statement
There are no conflicts to declare.
Figures

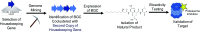





References
-
- Demain A. L. Fang A. Adv. Biochem. Eng./Biotechnol. 2000;132:1–39. - PubMed
-
- Atanasov A. G. Zotchev S. B. Dirsch V. M. Orhan I. E. Banach M. Rollinger J. M. Barreca D. Weckwerth W. Bauer R. Bayer E. A. Majeed M. Bishayee A. Bochkov V. Bonn G. K. Braidy N. Bucar F. Cifuentes A. D'Onofrio G. Bodkin M. Diederich M. Dinkova-Kostova A. T. Efferth T. El Bairi K. Arkells N. Fan T. P. Fiebich B. L. Freissmuth M. Georgiev M. I. Gibbons S. Godfrey K. M. Gruber C. W. Heer J. Huber L. A. Ibanez E. Kijjoa A. Kiss A. K. Lu A. Macias F. A. Miller M. J. S. Mocan A. Müller R. Nicoletti F. Perry G. Pittalà V. Rastrelli L. Ristow M. Russo G. L. Silva A. S. Schuster D. Sheridan H. Skalicka-Woźniak K. Skaltsounis L. Sobarzo-Sánchez E. Bredt D. S. Stuppner H. Sureda A. Tzvetkov N. T. Vacca R. A. Aggarwal B. B. Battino M. Giampieri F. Wink M. Wolfender J. L. Xiao J. Yeung A. W. K. Lizard G. Popp M. A. Heinrich M. Berindan-Neagoe I. Stadler M. Daglia M. Verpoorte R. Supuran C. T. Nat. Rev. Drug Discovery. 2021;20:200–216. doi: 10.1038/s41573-020-00114-z. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

