Machine Learning of Reaction Properties via Learned Representations of the Condensed Graph of Reaction
- PMID: 34734699
- PMCID: PMC9092344
- DOI: 10.1021/acs.jcim.1c00975
Machine Learning of Reaction Properties via Learned Representations of the Condensed Graph of Reaction
Abstract
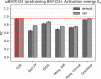
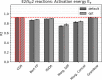
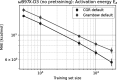
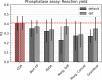
The estimation of chemical reaction properties such as activation energies, rates, or yields is a central topic of computational chemistry. In contrast to molecular properties, where machine learning approaches such as graph convolutional neural networks (GCNNs) have excelled for a wide variety of tasks, no general and transferable adaptations of GCNNs for reactions have been developed yet. We therefore combined a popular cheminformatics reaction representation, the so-called condensed graph of reaction (CGR), with a recent GCNN architecture to arrive at a versatile, robust, and compact deep learning model. The CGR is a superposition of the reactant and product graphs of a chemical reaction and thus an ideal input for graph-based machine learning approaches. The model learns to create a data-driven, task-dependent reaction embedding that does not rely on expert knowledge, similar to current molecular GCNNs. Our approach outperforms current state-of-the-art models in accuracy, is applicable even to imbalanced reactions, and possesses excellent predictive capabilities for diverse target properties, such as activation energies, reaction enthalpies, rate constants, yields, or reaction classes. We furthermore curated a large set of atom-mapped reactions along with their target properties, which can serve as benchmark data sets for future work. All data sets and the developed reaction GCNN model are available online, free of charge, and open source.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Gilmer J.; Schoenholz S. S.; Riley P. F.; Vinyals O.; Dahl G. E.. Neural Message Passing for Quantum Chemistry. In International Conference on Machine Learning, 2017; pp 1263–1272.
-
- Klicpera J.; Groß J.; Günnemann S.. Directional Message Passing for Molecular Graphs. arXiv preprint, arXiv:2003.03123, 2020.
-
- Zhang S.; Liu Y.; Xie L.. Molecular Mechanics-Driven Graph Neural Network with Multiplex Graph for Molecular Structures. arXiv preprint, arXiv:2011.07457, 2020.
-
- Alperstein Z.; Cherkasov A.; Rolfe J. T.. All Smiles Variational Autoencoder. arXiv preprint, arXiv:1905.13343, 2019.
-
- Zaslavskiy M.; Jégou S.; Tramel E. W.; Wainrib G. ToxicBlend: Virtual Screening of Toxic Compounds with Ensemble Predictors. Comp. Toxicol. 2019, 10, 81–88. 10.1016/j.comtox.2019.01.001. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

