The chromatin insulator CTCF regulates HPV18 transcript splicing and differentiation-dependent late gene expression
- PMID: 34735550
- PMCID: PMC8594839
- DOI: 10.1371/journal.ppat.1010032
The chromatin insulator CTCF regulates HPV18 transcript splicing and differentiation-dependent late gene expression
Abstract
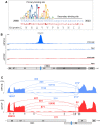
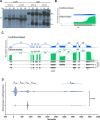
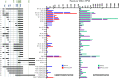
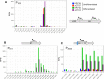
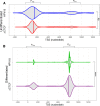
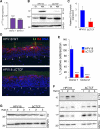
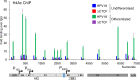
The ubiquitous host protein, CCCTC-binding factor (CTCF), is an essential regulator of cellular transcription and functions to maintain epigenetic boundaries, stabilise chromatin loops and regulate splicing of alternative exons. We have previously demonstrated that CTCF binds to the E2 open reading frame (ORF) of human papillomavirus (HPV) 18 and functions to repress viral oncogene expression in undifferentiated keratinocytes by co-ordinating an epigenetically repressed chromatin loop within HPV episomes. Keratinocyte differentiation disrupts CTCF-dependent chromatin looping of HPV18 episomes promoting induction of enhanced viral oncogene expression. To further characterise CTCF function in HPV transcription control we utilised direct, long-read Nanopore RNA-sequencing which provides information on the structure and abundance of full-length transcripts. Nanopore analysis of primary human keratinocytes containing HPV18 episomes before and after synchronous differentiation allowed quantification of viral transcript species, including the identification of low abundance novel transcripts. Comparison of transcripts produced in wild type HPV18 genome-containing cells to those identified in CTCF-binding deficient genome-containing cells identifies CTCF as a key regulator of differentiation-dependent late promoter activation, required for efficient E1^E4 and L1 protein expression. Furthermore, our data show that CTCF binding at the E2 ORF promotes usage of the downstream weak splice donor (SD) sites SD3165 and SD3284, to the dominant E4 splice acceptor site at nucleotide 3434. These findings demonstrate that in the HPV life cycle both early and late virus transcription programmes are facilitated by recruitment of CTCF to the E2 ORF.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Ruesch MN, Stubenrauch F, Laimins LA. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72(6):5016–24. doi: 10.1128/JVI.72.6.5016-5024.1998 ; PubMed Central PMCID: PMC110064. - DOI - PMC - PubMed
-
- Wooldridge TR, Laimins LA. Regulation of human papillomavirus type 31 gene expression during the differentiation-dependent life cycle through histone modifications and transcription factor binding. Virology. 2008;374(2):371–80. doi: 10.1016/j.virol.2007.12.011 ; PubMed Central PMCID: PMC2410142. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

