Constitutive activation of the PI3K-Akt-mTORC1 pathway sustains the m.3243 A > G mtDNA mutation
- PMID: 34737295
- PMCID: PMC8568893
- DOI: 10.1038/s41467-021-26746-2
Constitutive activation of the PI3K-Akt-mTORC1 pathway sustains the m.3243 A > G mtDNA mutation
Abstract
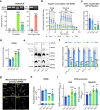
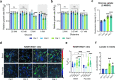
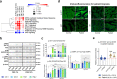
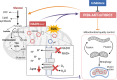
Mutations of the mitochondrial genome (mtDNA) cause a range of profoundly debilitating clinical conditions for which treatment options are very limited. Most mtDNA diseases show heteroplasmy - tissues express both wild-type and mutant mtDNA. While the level of heteroplasmy broadly correlates with disease severity, the relationships between specific mtDNA mutations, heteroplasmy, disease phenotype and severity are poorly understood. We have carried out extensive bioenergetic, metabolomic and RNAseq studies on heteroplasmic patient-derived cells carrying the most prevalent disease related mtDNA mutation, the m.3243 A > G. These studies reveal that the mutation promotes changes in metabolites which are associated with the upregulation of the PI3K-Akt-mTORC1 axis in patient-derived cells and tissues. Remarkably, pharmacological inhibition of PI3K, Akt, or mTORC1 reduced mtDNA mutant load and partially rescued cellular bioenergetic function. The PI3K-Akt-mTORC1 axis thus represents a potential therapeutic target that may benefit people suffering from the consequences of the m.3243 A > G mutation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

