Tubulin isotypes optimize distinct spindle positioning mechanisms during yeast mitosis
- PMID: 34739032
- PMCID: PMC8576917
- DOI: 10.1083/jcb.202010155
Tubulin isotypes optimize distinct spindle positioning mechanisms during yeast mitosis
Abstract
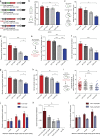
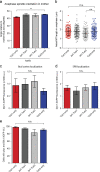
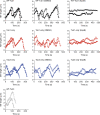
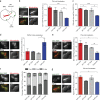
Microtubules are dynamic cytoskeleton filaments that are essential for a wide range of cellular processes. They are polymerized from tubulin, a heterodimer of α- and β-subunits. Most eukaryotic organisms express multiple isotypes of α- and β-tubulin, yet their functional relevance in any organism remains largely obscure. The two α-tubulin isotypes in budding yeast, Tub1 and Tub3, are proposed to be functionally interchangeable, yet their individual functions have not been rigorously interrogated. Here, we develop otherwise isogenic yeast strains expressing single tubulin isotypes at levels comparable to total tubulin in WT cells. Using genome-wide screening, we uncover unique interactions between the isotypes and the two major mitotic spindle positioning mechanisms. We further exploit these cells to demonstrate that Tub1 and Tub3 optimize spindle positioning by differentially recruiting key components of the Dyn1- and Kar9-dependent mechanisms, respectively. Our results provide novel mechanistic insights into how tubulin isotypes allow highly conserved microtubules to function in diverse cellular processes.
© 2021 Nsamba et al.
Figures



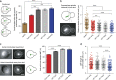
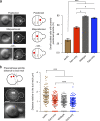
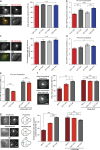

Comment in
-
Specialist α-tubulins for pluralist microtubules.J Cell Biol. 2021 Dec 6;220(12):e202110038. doi: 10.1083/jcb.202110038. Epub 2021 Nov 11. J Cell Biol. 2021. PMID: 34762120 Free PMC article.
References
-
- Bahi-Buisson, N., Poirier K., Fourniol F., Saillour Y., Valence S., Lebrun N., Hully M., Bianco C.F., Boddaert N., Elie C., et al. . LIS-Tubulinopathies Consortium . 2014. The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain. 137:1676–1700. 10.1093/brain/awu082 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

