GNS561, a clinical-stage PPT1 inhibitor, is efficient against hepatocellular carcinoma via modulation of lysosomal functions
- PMID: 34740311
- PMCID: PMC9037544
- DOI: 10.1080/15548627.2021.1988357
GNS561, a clinical-stage PPT1 inhibitor, is efficient against hepatocellular carcinoma via modulation of lysosomal functions
Abstract
Hepatocellular carcinoma is the most frequent primary liver cancer. Macroautophagy/autophagy inhibitors have been extensively studied in cancer but, to date, none has reached efficacy in clinical trials. In this study, we demonstrated that GNS561, a new autophagy inhibitor, whose anticancer activity was previously linked to lysosomal cell death, displayed high liver tropism and potent antitumor activity against a panel of human cancer cell lines and in two hepatocellular carcinoma in vivo models. We showed that due to its lysosomotropic properties, GNS561 could reach and specifically inhibited its enzyme target, PPT1 (palmitoyl-protein thioesterase 1), resulting in lysosomal unbound Zn2+ accumulation, impairment of cathepsin activity, blockage of autophagic flux, altered location of MTOR (mechanistic target of rapamycin kinase), lysosomal membrane permeabilization, caspase activation and cell death. Accordingly, GNS561, for which a global phase 1b clinical trial in liver cancers was just successfully achieved, represents a promising new drug candidate and a hopeful therapeutic strategy in cancer treatment.Abbreviations: ANXA5:annexin A5; ATCC: American type culture collection; BafA1: bafilomycin A1; BSA: bovine serum albumin; CASP3: caspase 3; CASP7: caspase 7; CASP8: caspase 8; CCND1: cyclin D1; CTSB: cathepsin B; CTSD: cathepsin D; CTSL: cathepsin L; CQ: chloroquine; iCCA: intrahepatic cholangiocarcinoma; DEN: diethylnitrosamine; DMEM: Dulbelcco's modified Eagle medium; FBS: fetal bovine serum; FITC: fluorescein isothiocyanate; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; HCC: hepatocellular carcinoma; HCQ: hydroxychloroquine; HDSF: hexadecylsulfonylfluoride; IC50: mean half-maximal inhibitory concentration; LAMP: lysosomal associated membrane protein; LC3-II: phosphatidylethanolamine-conjugated form of MAP1LC3; LMP: lysosomal membrane permeabilization; MALDI: matrix assisted laser desorption ionization; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MKI67: marker of proliferation Ki-67; MTOR: mechanistic target of rapamycin kinase; MRI: magnetic resonance imaging; NH4Cl: ammonium chloride; NtBuHA: N-tert-butylhydroxylamine; PARP: poly(ADP-ribose) polymerase; PBS: phosphate-buffered saline; PPT1: palmitoyl-protein thioesterase 1; SD: standard deviation; SEM: standard error mean; vs, versus; Zn2+: zinc ion; Z-Phe: Z-Phe-Tyt(tBu)-diazomethylketone; Z-VAD-FMK: carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]- fluoromethylketone.
Keywords: Antitumor; PPT1; autophagy; liver cancer; lysosome; mtor.
Conflict of interest statement
SB, EB, FB, ER, SM, MR, MN, JT, JC, CD, GJ, CS, CA and PH are employees of Genoscience Pharma. SB, ER, FB, CD, CS, CA and PH are shareholders of Genoscience Pharma. SB, FB, JC and PH are co-inventors of a pending patent. The other authors declare that they have no conflicts of interest to report.
Figures

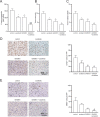
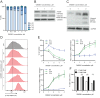
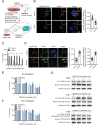
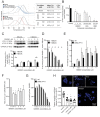
References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Aits S, Jäättelä M.. Lysosomal cell death at a glance. J Cell Sci. 2013;126(9):1905–1912. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
