Role of mutations and post-translational modifications in systemic AL amyloidosis studied by cryo-EM
- PMID: 34741031
- PMCID: PMC8571268
- DOI: 10.1038/s41467-021-26553-9
Role of mutations and post-translational modifications in systemic AL amyloidosis studied by cryo-EM
Abstract
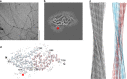
Systemic AL amyloidosis is a rare disease that is caused by the misfolding of immunoglobulin light chains (LCs). Potential drivers of amyloid formation in this disease are post-translational modifications (PTMs) and the mutational changes that are inserted into the LCs by somatic hypermutation. Here we present the cryo electron microscopy (cryo-EM) structure of an ex vivo λ1-AL amyloid fibril whose deposits disrupt the ordered cardiomyocyte structure in the heart. The fibril protein contains six mutational changes compared to the germ line and three PTMs (disulfide bond, N-glycosylation and pyroglutamylation). Our data imply that the disulfide bond, glycosylation and mutational changes contribute to determining the fibril protein fold and help to generate a fibril morphology that is able to withstand proteolytic degradation inside the body.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Merlini G, et al. Systemic immunoglobulin light chain amyloidosis. Nat. Rev. Dis. Prim. 2018;4:1–19. - PubMed
-
- Janeway C. A. Jr., Travers, P., Walport, M. & Shlomchik, M. J. Immunobiology: the Immune System in Health and Disease. 5th edn (Garland Science, 2001). The generation of diversity in immunoglobulins.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

