The tumor suppressor MIR139 is silenced by POLR2M to promote AML oncogenesis
- PMID: 34741119
- PMCID: PMC8885418
- DOI: 10.1038/s41375-021-01461-5
The tumor suppressor MIR139 is silenced by POLR2M to promote AML oncogenesis
Abstract
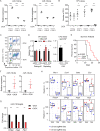
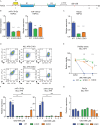
MIR139 is a tumor suppressor and is commonly silenced in acute myeloid leukemia (AML). However, the tumor-suppressing activities of miR-139 and molecular mechanisms of MIR139-silencing remain largely unknown. Here, we studied the poorly prognostic MLL-AF9 fusion protein-expressing AML. We show that MLL-AF9 expression in hematopoietic precursors caused epigenetic silencing of MIR139, whereas overexpression of MIR139 inhibited in vitro and in vivo AML outgrowth. We identified novel miR-139 targets that mediate the tumor-suppressing activities of miR-139 in MLL-AF9 AML. We revealed that two enhancer regions control MIR139 expression and found that the polycomb repressive complex 2 (PRC2) downstream of MLL-AF9 epigenetically silenced MIR139 in AML. Finally, a genome-wide CRISPR-Cas9 knockout screen revealed RNA Polymerase 2 Subunit M (POLR2M) as a novel MIR139-regulatory factor. Our findings elucidate the molecular control of tumor suppressor MIR139 and reveal a role for POLR2M in the MIR139-silencing mechanism, downstream of MLL-AF9 and PRC2 in AML. In addition, we confirmed these findings in human AML cell lines with different oncogenic aberrations, suggesting that this is a more common oncogenic mechanism in AML. Our results may pave the way for new targeted therapy in AML.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Steinhilber D, Marschalek R. How to effectively treat acute leukemia patients bearing MLL-rearrangements? Biochem Pharm. 2018;147:183–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

