Comparative Study of the Gut Microbiota Among Four Different Marine Mammals in an Aquarium
- PMID: 34745077
- PMCID: PMC8567075
- DOI: 10.3389/fmicb.2021.769012
Comparative Study of the Gut Microbiota Among Four Different Marine Mammals in an Aquarium
Abstract
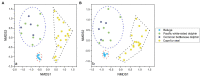
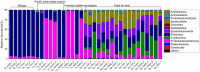
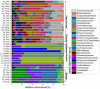
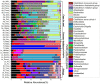
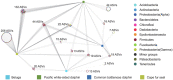
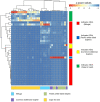
Despite an increasing appreciation in the importance of host-microbe interactions in ecological and evolutionary processes, information on the gut microbial communities of some marine mammals is still lacking. Moreover, whether diet, environment, or host phylogeny has the greatest impact on microbial community structure is still unknown. To fill part of this knowledge gap, we exploited a natural experiment provided by an aquarium with belugas (Delphinapterus leucas) affiliated with family Monodontidae, Pacific white-sided dolphins (Lagenorhynchus obliquidens) and common bottlenose dolphin (Tursiops truncatus) affiliated with family Delphinidae, and Cape fur seals (Arctocephalus pusillus pusillus) affiliated with family Otariidae. Results show significant differences in microbial community composition of whales, dolphins, and fur seals and indicate that host phylogeny (family level) plays the most important role in shaping the microbial communities, rather than food and environment. In general, the gut microbial communities of dolphins had significantly lower diversity compared to that of whales and fur seals. Overall, the gut microbial communities were mainly composed of Firmicutes and Gammaproteobacteria, together with some from Bacteroidetes, Fusobacteria, and Epsilonbacteraeota. However, specific bacterial lineages were differentially distributed among the marine mammal groups. For instance, Lachnospiraceae, Ruminococcaceae, and Peptostreptococcaceae were the dominant bacterial lineages in the gut of belugas, while for Cape fur seals, Moraxellaceae and Bacteroidaceae were the main bacterial lineages. Moreover, gut microbial communities in both Pacific white-sided dolphins and common bottlenose dolphins were dominated by a number of pathogenic bacteria, including Clostridium perfringens, Vibrio fluvialis, and Morganella morganii, reflecting the poor health condition of these animals. Although there is a growing recognition of the role microorganisms play in the gut of marine mammals, current knowledge about these microbial communities is still severely lacking. Large-scale research studies should be undertaken to reveal the roles played by the gut microbiota of different marine mammal species.
Keywords: Cape fur seal; Pacific white-sided dolphin; beluga; common bottlenose dolphin; gut microbial communities.
Copyright © 2021 Bai, Zhang, Zhang, Du, Du, Zhu, Liu, Xie and Li.
Conflict of interest statement
CZ and XD are employed by Atlantis Sanya. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bai S., Zhang P., Lin M., Lin W., Yang Z., Li S. (2021). Microbial diversity and structure in the gastrointestinal tracts of two stranded short-finned pilot whales (Globicephala macrorhynchus) and a pygmy sperm whale (Kogia breviceps). Integr. Zool. 16, 324–335. doi: 10.1111/1749-4877.12502, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

