Unique Terminal Regions and Specific Deletions of the Segmented Double-Stranded RNA Genome of Alternaria Alternata Virus 1, in the Proposed Family Alternaviridae
- PMID: 34745080
- PMCID: PMC8570381
- DOI: 10.3389/fmicb.2021.773062
Unique Terminal Regions and Specific Deletions of the Segmented Double-Stranded RNA Genome of Alternaria Alternata Virus 1, in the Proposed Family Alternaviridae
Abstract
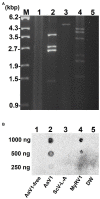
Alternaria alternata virus 1 (AaV1) has been identified in the saprophytic fungus Alternaria alternata strain EGS 35-193. AaV1 has four genomic double-stranded (ds)RNA segments (dsRNA1-4) packaged in isometric particles. The 3' end of each coding strand is polyadenylated (36-50nt), but the presence of a cap structure at each 5' end has not previously been investigated. Here, we have characterized the AaV1 genome and found that it has unique features among the mycoviruses. We confirmed the existence of cap structures on the 5' ends of the AaV1 genomic dsRNAs using RNA dot blots with anti-cap antibodies and the oligo-capping method. Polyclonal antibodies against purified AaV1 particles specifically bound to an 82kDa protein, suggesting that this protein is the major capsid component. Subsequent Edman degradation indicated that the AaV1 dsRNA3 segment encodes the major coat protein. Two kinds of defective AaV1 dsRNA2, which is 2,794bp (844 aa) in length when intact, appeared in EGS 35-193 during subculturing, as confirmed by RT-PCR and northern hybridization. Sequence analysis revealed that one of the two defective dsRNA2s contained a 231bp deletion, while the other carried both the 231bp deletion and an additional 465bp deletion in the open reading frame. Both deletions occurred in-frame, resulting in predicted proteins of 767 aa and 612 aa. The fungal isolates carrying virions with the defective dsRNA2s showed impaired growth and abnormal pigmentation. To our best knowledge, AaV1 is the first dsRNA virus to be identified with both 5' cap and 3'poly(A) structures on its genomic segments, as well as the specific deletions of dsRNA2.
Keywords: 5' cap structure; Alternaria alternata; Alternaviridae; deletion of RNA genome; dsRNA virus; mycovirus; viral protein.
Copyright © 2021 Wu, Aoki, Takeshita, Fukuhara, Chiura, Arie, Kotta-Loizou, Okada, Komatsu and Moriyama.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., et al. . (2009). Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. U. S. A. 106, 8157–8162. doi: 10.1073/pnas.0901931106 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

