Mitochondrial Fission Factor Is a Novel Interacting Protein of the Critical B Cell Survival Regulator TRAF3 in B Lymphocytes
- PMID: 34745083
- PMCID: PMC8564014
- DOI: 10.3389/fimmu.2021.670338
Mitochondrial Fission Factor Is a Novel Interacting Protein of the Critical B Cell Survival Regulator TRAF3 in B Lymphocytes
Abstract
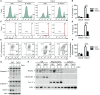
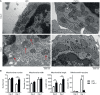
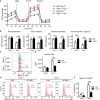
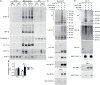
Proteins controlling mitochondrial fission have been recognized as essential regulators of mitochondrial functions, mitochondrial quality control and cell apoptosis. In the present study, we identified the critical B cell survival regulator TRAF3 as a novel binding partner of the key mitochondrial fission factor, MFF, in B lymphocytes. Elicited by our unexpected finding that the majority of cytoplasmic TRAF3 proteins were localized at the mitochondria in resting splenic B cells after ex vivo culture for 2 days, we found that TRAF3 specifically interacted with MFF as demonstrated by co-immunoprecipitation and GST pull-down assays. We further found that in the absence of stimulation, increased protein levels of mitochondrial TRAF3 were associated with altered mitochondrial morphology, decreased mitochondrial respiration, increased mitochondrial ROS production and membrane permeabilization, which eventually culminated in mitochondria-dependent apoptosis in resting B cells. Loss of TRAF3 had the opposite effects on the morphology and function of mitochondria as well as mitochondria-dependent apoptosis in resting B cells. Interestingly, co-expression of TRAF3 and MFF resulted in decreased phosphorylation and ubiquitination of MFF as well as decreased ubiquitination of TRAF3. Moreover, lentivirus-mediated overexpression of MFF restored mitochondria-dependent apoptosis in TRAF3-deficient malignant B cells. Taken together, our findings provide novel insights into the apoptosis-inducing mechanisms of TRAF3 in B cells: as a result of survival factor deprivation or under other types of stress, TRAF3 is mobilized to the mitochondria through its interaction with MFF, where it triggers mitochondria-dependent apoptosis. This new role of TRAF3 in controlling mitochondrial homeostasis might have key implications in TRAF3-mediated regulation of B cell transformation in different cellular contexts. Our findings also suggest that mitochondrial fission is an actionable therapeutic target in human B cell malignancies, including those with TRAF3 deletion or relevant mutations.
Keywords: B cell malignancies; B lymphocytes; MFF; TRAF3; apoptosis; mitochondria.
Copyright © 2021 Liu, Gokhale, Jung, Zhu, Luo, Saha, Guo, Zhang, Kyin, Zong, White and Xie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ruddon R. The Epidemiology of Human Cancer. In: Ruddon RW, editor. Cancer Biology, 4th ed. Oxford, England: Oxford University Press; (2007). p. 62–116.
-
- Horner MJ, Ries LAG, Krapcho M, Neyman N, Aminou R, Howlader N, et al. . Surveillance, Epidemiology, and End Results (SEER) Program. SEER Cancer Statistics Review, 1975-2006. Bethesda, MD: National Cancer Institute; (2008). Available at: www.seer.cancer.gov. (Nov 2008 Sub).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

