Transcriptomic Analysis Identifies A Tolerogenic Dendritic Cell Signature
- PMID: 34745103
- PMCID: PMC8564488
- DOI: 10.3389/fimmu.2021.733231
Transcriptomic Analysis Identifies A Tolerogenic Dendritic Cell Signature
Abstract
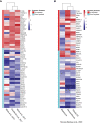
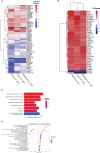
Dendritic cells (DC) are central to regulating innate and adaptive immune responses. Strategies that modify DC function provide new therapeutic opportunities in autoimmune diseases and transplantation. Current pharmacological approaches can alter DC phenotype to induce tolerogenic DC (tolDC), a maturation-resistant DC subset capable of directing a regulatory immune response that are being explored in current clinical trials. The classical phenotypic characterization of tolDC is limited to cell-surface marker expression and anti-inflammatory cytokine production, although these are not specific. TolDC may be better defined using gene signatures, but there is no consensus definition regarding genotypic markers. We address this shortcoming by analyzing available transcriptomic data to yield an independent set of differentially expressed genes that characterize human tolDC. We validate this transcriptomic signature and also explore gene differences according to the method of tolDC generation. As well as establishing a novel characterization of tolDC, we interrogated its translational utility in vivo, demonstrating this geneset was enriched in the liver, a known tolerogenic organ. Our gene signature will potentially provide greater understanding regarding transcriptional regulators of tolerance and allow researchers to standardize identification of tolDC used for cellular therapy in clinical trials.
Keywords: dendritic cell; gene expression profile analysis; human dendritic cell; liver; mature dendritic cells; mononuclear phagocyte cells; tolerogenic dendritic cell (tolDC); transcriptomic.
Copyright © 2021 Robertson, Li, Kim, Rhodes, Harman, Patrick and Rogers.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

