Functional Genomics of PRUNE1 in Neurodevelopmental Disorders (NDDs) Tied to Medulloblastoma (MB) and Other Tumors
- PMID: 34745995
- PMCID: PMC8569853
- DOI: 10.3389/fonc.2021.758146
Functional Genomics of PRUNE1 in Neurodevelopmental Disorders (NDDs) Tied to Medulloblastoma (MB) and Other Tumors
Abstract
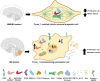
We analyze the fundamental functions of Prune_1 in brain pathophysiology. We discuss the importance and maintenance of the function of Prune_1 and how its perturbation influences both brain pathological conditions, neurodevelopmental disorder with microcephaly, hypotonia, and variable brain anomalies (NMIHBA; OMIM: 617481), and tumorigenesis of medulloblastoma (MB) with functional correlations to other tumors. A therapeutic view underlying recent discoveries identified small molecules and cell penetrating peptides to impair the interaction of Prune_1 with protein partners (e.g., Nm23-H1), thus further impairing intracellular and extracellular signaling (i.e., canonical Wnt and TGF-β pathways). Identifying the mechanism of action of Prune_1 as responsible for neurodevelopmental disorders (NDDs), we have recognized other genes which are found overexpressed in brain tumors (e.g., MB) with functional implications in neurodevelopmental processes, as mainly linked to changes in mitotic cell cycle processes. Thus, with Prune_1 being a significant target in NDDs, we discuss how its network of action can be dysregulated during brain development, thus generating cancer and metastatic dissemination.
Keywords: PRUNE_1; TGF—transforming growth factor; Wnt; medulloblastoma; metastasis; microtubules polymerization; neurodevelopmental disorder with microcephaly, hypotonia, variable brain anomalies (NMIHBA); proliferation rate.
Copyright © 2021 Bibbò, Sorice, Ferrucci and Zollo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

