Epigenetic signature of chronic low back pain in human T cells
- PMID: 34746619
- PMCID: PMC8568391
- DOI: 10.1097/PR9.0000000000000960
Epigenetic signature of chronic low back pain in human T cells
Abstract
Objective: Determine if chronic low back pain (LBP) is associated with DNA methylation signatures in human T cells that will reveal novel mechanisms and potential therapeutic targets and explore the feasibility of epigenetic diagnostic markers for pain-related pathophysiology.
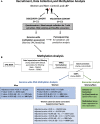
Methods: Genome-wide DNA methylation analysis of 850,000 CpG sites in women and men with chronic LBP and pain-free controls was performed. T cells were isolated (discovery cohort, n = 32) and used to identify differentially methylated CpG sites, and gene ontologies and molecular pathways were identified. A polygenic DNA methylation score for LBP was generated in both women and men. Validation was performed in an independent cohort (validation cohort, n = 63) of chronic LBP and healthy controls.
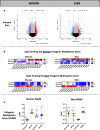
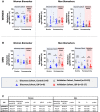
Results: Analysis with the discovery cohort revealed a total of 2,496 and 419 differentially methylated CpGs in women and men, respectively. In women, most of these sites were hypomethylated and enriched in genes with functions in the extracellular matrix, in the immune system (ie, cytokines), or in epigenetic processes. In men, a unique chronic LBP DNA methylation signature was identified characterized by significant enrichment for genes from the major histocompatibility complex. Sex-specific polygenic DNA methylation scores were generated to estimate the pain status of each individual and confirmed in the validation cohort using pyrosequencing.
Conclusion: This study reveals sex-specific DNA methylation signatures in human T cells that discriminates chronic LBP participants from healthy controls.
Keywords: Biomarker; DNA methylation; Epigenetics; Low back pain; T cells.
Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of The International Association for the Study of Pain.
Conflict of interest statement
Authors S. Gregoire, D. Cheishvili, M. Szyf, and L.S. Stone are coinventors on a provisional patent protecting the methylation signatures described here for possible use as a biomarker for chronic pain. D. Cheishvilli and M. Szyf are associated with HKG Epitherapeutics that develops epigenetic biomarkers for clinical use. The remaining authors have no conflicts of interest to declare.Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Figures


References
-
- Balagué F, Mannion AF, Pellisé F, Cedraschi C. Non-specific low back pain. Lancet 2012;379:482–91. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
