An integrated multi-omic analysis of iPSC-derived motor neurons from C9ORF72 ALS patients
- PMID: 34746695
- PMCID: PMC8554488
- DOI: 10.1016/j.isci.2021.103221
An integrated multi-omic analysis of iPSC-derived motor neurons from C9ORF72 ALS patients
Abstract
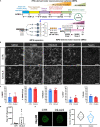
Neurodegenerative diseases are challenging for systems biology because of the lack of reliable animal models or patient samples at early disease stages. Induced pluripotent stem cells (iPSCs) could address these challenges. We investigated DNA, RNA, epigenetics, and proteins in iPSC-derived motor neurons from patients with ALS carrying hexanucleotide expansions in C9ORF72. Using integrative computational methods combining all omics datasets, we identified novel and known dysregulated pathways. We used a C9ORF72 Drosophila model to distinguish pathways contributing to disease phenotypes from compensatory ones and confirmed alterations in some pathways in postmortem spinal cord tissue of patients with ALS. A different differentiation protocol was used to derive a separate set of C9ORF72 and control motor neurons. Many individual -omics differed by protocol, but some core dysregulated pathways were consistent. This strategy of analyzing patient-specific neurons provides disease-related outcomes with small numbers of heterogeneous lines and reduces variation from single-omics to elucidate network-based signatures.
Keywords: Biological sciences; Neuroscience; Omics; Systems biology; Systems neuroscience.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

