Natural variation in the transcriptional response of Drosophila melanogaster to oxidative stress
- PMID: 34747443
- PMCID: PMC8727983
- DOI: 10.1093/g3journal/jkab366
Natural variation in the transcriptional response of Drosophila melanogaster to oxidative stress
Abstract
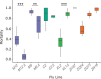
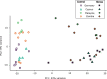
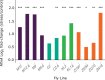
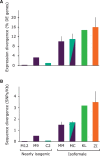
Broadly distributed species must cope with diverse and changing environmental conditions, including various forms of stress. Cosmopolitan populations of Drosophila melanogaster are more tolerant to oxidative stress than those from the species' ancestral range in sub-Saharan Africa, and the degree of tolerance is associated with an insertion/deletion polymorphism in the 3' untranslated region of the Metallothionein A (MtnA) gene that varies clinally in frequency. We examined oxidative stress tolerance and the transcriptional response to oxidative stress in cosmopolitan and sub-Saharan African populations of D. melanogaster, including paired samples with allelic differences at the MtnA locus. We found that the effect of the MtnA polymorphism on oxidative stress tolerance was dependent on the genomic background, with the deletion allele increasing tolerance only in a northern, temperate population. Genes that were differentially expressed under oxidative stress included MtnA and other metallothioneins, as well as those involved in glutathione metabolism and other genes known to be part of the oxidative stress response or the general stress response. A gene coexpression analysis revealed further genes and pathways that respond to oxidative stress including those involved in additional metabolic processes, autophagy, and apoptosis. There was a significant overlap among the genes induced by oxidative and cold stress, which suggests a shared response pathway to these two stresses. Interestingly, the MtnA deletion was associated with consistent changes in the expression of many genes across all genomic backgrounds, regardless of the expression level of the MtnA gene itself. We hypothesize that this is an indirect effect driven by the loss of microRNA binding sites within the MtnA 3' untranslated region.
Keywords: adaptation; gene expression; metallothionein; population genetics; transcriptomics.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


References
-
- Abel J, de Ruiter N.. 1989. Inhibition of hydroxyl-radical-generated DNA degradation by metallothionein. Toxicol Lett. 47:191–196. doi:10.1016/0378–4274(89)90075-1. - PubMed
-
- Akman SA, Dietrich M, Chlebowski R, Limberg P, Block JB.. 1985. Modulation of cytotoxicity of menadione sodium bisulfite versus leukemia L1210 by the acid-soluble thiol pool. Cancer Res. 45:5257–5262. - PubMed
-
- Atanesyan L, Günther V, Celniker SE, Georgiev O, Schaffner W.. 2011. Characterization of MtnE, the fifth metallothionein member in Drosophila. J Biol Inorg Chem. 16:1047–1056. doi:10.1007/s00775-011–0825-4. - PubMed
-
- Baker H, DeAngelis B, Frank O, Khalil M, Hutner SH, et al.1996. Antioxidant survey to assess antagonism to redox stress using a prokaryotic and an eukaryotic system. Experientia. 52:597–599. doi:10.1007/BF01969736. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
