PLAZA 5.0: extending the scope and power of comparative and functional genomics in plants
- PMID: 34747486
- PMCID: PMC8728282
- DOI: 10.1093/nar/gkab1024
PLAZA 5.0: extending the scope and power of comparative and functional genomics in plants
Abstract
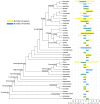
PLAZA is a platform for comparative, evolutionary, and functional plant genomics. It makes a broad set of genomes, data types and analysis tools available to researchers through a user-friendly website, an API, and bulk downloads. In this latest release of the PLAZA platform, we are integrating a record number of 134 high-quality plant genomes, split up over two instances: PLAZA Dicots 5.0 and PLAZA Monocots 5.0. This number of genomes corresponds with a massive expansion in the number of available species when compared to PLAZA 4.0, which offered access to 71 species, a 89% overall increase. The PLAZA 5.0 release contains information for 5 882 730 genes, and offers pre-computed gene families and phylogenetic trees for 5 274 684 protein-coding genes. This latest release also comes with a set of new and updated features: a new BED import functionality for the workbench, improved interactive visualizations for functional enrichments and genome-wide mapping of gene sets, and a fully redesigned and extended API. Taken together, this new version offers extended support for plant biologists working on different families within the green plant lineage and provides an efficient and versatile toolbox for plant genomics. All PLAZA releases are accessible from the portal website: https://bioinformatics.psb.ugent.be/plaza/.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Jung H., Winefield C., Bombarely A., Prentis P., Waterhouse P.. Tools and strategies for long-read sequencing and de novo assembly of plant genomes. Trends Plant Sci. 2019; 24:700–724. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

