Resolving Dynamics in the Ensemble: Finding Paths through Intermediate States and Disordered Protein Structures
- PMID: 34748336
- PMCID: PMC9096987
- DOI: 10.1021/acs.jpcb.1c05820
Resolving Dynamics in the Ensemble: Finding Paths through Intermediate States and Disordered Protein Structures
Abstract
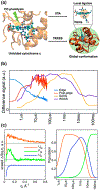
Proteins have been found to inhabit a diverse set of three-dimensional structures. The dynamics that govern protein interconversion between structures happen over a wide range of time scales─picoseconds to seconds. Our understanding of protein functions and dynamics is largely reliant upon our ability to elucidate physically populated structures. From an experimental structural characterization perspective, we are often limited to measuring the ensemble-averaged structure both in the steady-state and time-resolved regimes. Generating kinetic models and understanding protein structure-function relationships require atomistic knowledge of the populated states in the ensemble. In this Perspective, we present ensemble refinement methodologies that integrate time-resolved experimental signals with molecular dynamics models. We first discuss integration of experimental structural restraints to molecular models in disordered protein systems that adhere to the principle of maximum entropy for creating a complete set of ensemble structures. We then propose strategies to find kinetic pathways between the refined structures, using time-resolved inputs to guide molecular dynamics trajectories and the use of inference to generate tailored stimuli to prepare a desired ensemble of protein states.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Liu Y; Wang X; Liu B A Comprehensive Review and Comparison of Existing Computational Methods for Intrinsically Disordered Protein and Region Prediction. Briefings Bioinf. 2019, 20, 330–346. - PubMed
-
- Michaelis M; Hildebrand N; Meißner RH; Wurzler N; Li Z; Hirst JD; Micsonai A; Kardos J; Delle Piane M; Colombi Ciacchi L Impact of the Conformational Variability of Oligopeptides on the Computational Prediction of Their Cd Spectra. J. Phys. Chem. B 2019, 123, 6694–6704. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

