GPRuler: Metabolic gene-protein-reaction rules automatic reconstruction
- PMID: 34748537
- PMCID: PMC8601613
- DOI: 10.1371/journal.pcbi.1009550
GPRuler: Metabolic gene-protein-reaction rules automatic reconstruction
Abstract
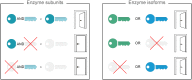
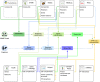
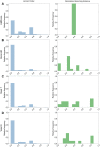
Metabolic network models are increasingly being used in health care and industry. As a consequence, many tools have been released to automate their reconstruction process de novo. In order to enable gene deletion simulations and integration of gene expression data, these networks must include gene-protein-reaction (GPR) rules, which describe with a Boolean logic relationships between the gene products (e.g., enzyme isoforms or subunits) associated with the catalysis of a given reaction. Nevertheless, the reconstruction of GPRs still remains a largely manual and time consuming process. Aiming at fully automating the reconstruction process of GPRs for any organism, we propose the open-source python-based framework GPRuler. By mining text and data from 9 different biological databases, GPRuler can reconstruct GPRs starting either from just the name of the target organism or from an existing metabolic model. The performance of the developed tool is evaluated at small-scale level for a manually curated metabolic model, and at genome-scale level for three metabolic models related to Homo sapiens and Saccharomyces cerevisiae organisms. By exploiting these models as benchmarks, the proposed tool shown its ability to reproduce the original GPR rules with a high level of accuracy. In all the tested scenarios, after a manual investigation of the mismatches between the rules proposed by GPRuler and the original ones, the proposed approach revealed to be in many cases more accurate than the original models. By complementing existing tools for metabolic network reconstruction with the possibility to reconstruct GPRs quickly and with a few resources, GPRuler paves the way to the study of context-specific metabolic networks, representing the active portion of the complete network in given conditions, for organisms of industrial or biomedical interest that have not been characterized metabolically yet.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Gunning PW. Protein isoforms and isozymes. eLS. 2005
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

