CCR2 Signaling Restricts SARS-CoV-2 Infection
- PMID: 34749524
- PMCID: PMC8576528
- DOI: 10.1128/mBio.02749-21
CCR2 Signaling Restricts SARS-CoV-2 Infection
Erratum in
-
Erratum for Vanderheiden et al., "CCR2 Signaling Restricts SARS-CoV-2 Infection".mBio. 2022 Jun 28;13(3):e0025922. doi: 10.1128/mbio.00259-22. Epub 2022 Apr 14. mBio. 2022. PMID: 35420471 Free PMC article. No abstract available.
Abstract
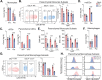
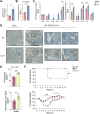
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a historic pandemic of respiratory disease (coronavirus disease 2019 [COVID-19]), and current evidence suggests that severe disease is associated with dysregulated immunity within the respiratory tract. However, the innate immune mechanisms that mediate protection during COVID-19 are not well defined. Here, we characterize a mouse model of SARS-CoV-2 infection and find that early CCR2 signaling restricts the viral burden in the lung. We find that a recently developed mouse-adapted SARS-CoV-2 (MA-SARS-CoV-2) strain as well as the emerging B.1.351 variant trigger an inflammatory response in the lung characterized by the expression of proinflammatory cytokines and interferon-stimulated genes. Using intravital antibody labeling, we demonstrate that MA-SARS-CoV-2 infection leads to increases in circulating monocytes and an influx of CD45+ cells into the lung parenchyma that is dominated by monocyte-derived cells. Single-cell RNA sequencing (scRNA-Seq) analysis of lung homogenates identified a hyperinflammatory monocyte profile. We utilize this model to demonstrate that mechanistically, CCR2 signaling promotes the infiltration of classical monocytes into the lung and the expansion of monocyte-derived cells. Parenchymal monocyte-derived cells appear to play a protective role against MA-SARS-CoV-2, as mice lacking CCR2 showed higher viral loads in the lungs, increased lung viral dissemination, and elevated inflammatory cytokine responses. These studies have identified a potential CCR2-monocyte axis that is critical for promoting viral control and restricting inflammation within the respiratory tract during SARS-CoV-2 infection. IMPORTANCE SARS-CoV-2 has caused a historic pandemic of respiratory disease (COVID-19), and current evidence suggests that severe disease is associated with dysregulated immunity within the respiratory tract. However, the innate immune mechanisms that mediate protection during COVID-19 are not well defined. Here, we characterize a mouse model of SARS-CoV-2 infection and find that early CCR2-dependent infiltration of monocytes restricts the viral burden in the lung. We find that SARS-CoV-2 triggers an inflammatory response in the lung characterized by the expression of proinflammatory cytokines and interferon-stimulated genes. Using RNA sequencing and flow cytometry approaches, we demonstrate that SARS-CoV-2 infection leads to increases in circulating monocytes and an influx of CD45+ cells into the lung parenchyma that is dominated by monocyte-derived cells. Mechanistically, CCR2 signaling promoted the infiltration of classical monocytes into the lung and the expansion of monocyte-derived cells. Parenchymal monocyte-derived cells appear to play a protective role against MA-SARS-CoV-2, as mice lacking CCR2 showed higher viral loads in the lungs, increased lung viral dissemination, and elevated inflammatory cytokine responses. These studies have identified that the CCR2 pathway is critical for promoting viral control and restricting inflammation within the respiratory tract during SARS-CoV-2 infection.
Keywords: SARS-CoV-2; innate immunity; lung inflammation; monocytes; mouse model.
Figures



Update of
-
CCR2-dependent monocyte-derived cells restrict SARS-CoV-2 infection.bioRxiv [Preprint]. 2021 May 4:2021.05.03.442538. doi: 10.1101/2021.05.03.442538. bioRxiv. 2021. Update in: mBio. 2021 Dec 21;12(6):e0274921. doi: 10.1128/mBio.02749-21. PMID: 33972938 Free PMC article. Updated. Preprint.
References
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Yin X, Riva L, Pu Y, Martin-Sancho L, Kanamune J, Yamamoto Y, Sakai K, Gotoh S, Miorin L, De Jesus PD, Yang C-C, Herbert KM, Yoh S, Hultquist JF, García-Sastre A, Chanda SK. 2021. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep 34:108628. doi:10.1016/j.celrep.2020.108628. - DOI - PMC - PubMed
-
- Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, Wagh D, Coller J, Pellegrini KL, Kazmin D, Alaaeddine G, Leung WS, Chan JMC, Chik TSH, Choi CYC, Huerta C, Paine McCullough M, Lv H, Anderson E, Edupuganti S, Upadhyay AA, Bosinger SE, Maecker HT, Khatri P, Rouphael N, Peiris M, Pulendran B. 2020. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369:1210–1220. doi:10.1126/science.abc6261. - DOI - PMC - PubMed
-
- Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, Aoued H, Tharp GM, Pellegrini KL, Manfredi C, Sorscher E, Mainou B, Lobby JL, Kohlmeier JE, Lowen AC, Shi P-Y, Menachery VD, Anderson LJ, Grakoui A, Bosinger SE, Suthar MS. 2020. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J Virol 94:e00985-20. doi:10.1128/JVI.00985-20. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI136533/AI/NIAID NIH HHS/United States
- R35 HL150803/HL/NHLBI NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- T32 AI074492/AI/NIAID NIH HHS/United States
- P51 OD011132/CD/ODCDC CDC HHS/United States
- S10 OD026799/OD/NIH HHS/United States
- HHSN272201400004C/AI/NIAID NIH HHS/United States
- Emory Executive Vice President for Health Affairs Synergy Award
- R56 AI147623/AI/NIAID NIH HHS/United States
- Emory-UGA Center of Excellence for Influenza Research and Surveillance
- Woodruff Health Science Center 2020 COVID-19 CURE Award
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
