Restraint of Fumarate Accrual by HIF-1α Preserves miR-27a-Mediated Limitation of Interleukin 10 during Infection of Macrophages by Histoplasma capsulatum
- PMID: 34749531
- PMCID: PMC8576535
- DOI: 10.1128/mBio.02710-21
Restraint of Fumarate Accrual by HIF-1α Preserves miR-27a-Mediated Limitation of Interleukin 10 during Infection of Macrophages by Histoplasma capsulatum
Abstract
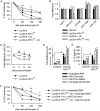
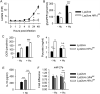
Hypoxia-inducible factor 1α (HIF-1α) regulates the immunometabolic phenotype of macrophages, including the orchestration of inflammatory and antimicrobial processes. Macrophages deficient in HIF-1α produce excessive quantities of the anti-inflammatory cytokine interleukin 10 (IL-10) during infection with the intracellular fungal pathogen Histoplasma capsulatum (R. A. Fecher, M. C. Horwath, D. Friedrich, J. Rupp, G. S. Deepe, J Immunol 197:565-579, 2016, https://doi.org/10.4049/jimmunol.1600342). Thus, the macrophage fails to become activated in response to proinflammatory cytokines and remains the intracellular niche of the pathogen. Here, we identify the tricarboxylic acid (TCA) cycle metabolite fumarate as the driver of IL-10 during macrophage infection with H. capsulatum in the absence of HIF-1α. Accumulation of fumarate reduced expression of a HIF-1α-dependent microRNA (miRNA), miR-27a, known to mediate decay of Il10 mRNA. Inhibition of fumarate accrual in vivo limited IL-10 and fungal growth. Our data demonstrate the critical role of HIF-1α in shaping appropriate TCA cycle activity in response to infection and highlight the consequences of a dysregulated immunometabolic response. IMPORTANCE Histoplasma capsulatum and related Histoplasma species are intracellular fungal pathogens endemic to broad regions of the globe, including the Americas, Africa, and Asia. While most infections resolve with mild or no symptoms, failure of the host to control fungal growth produces severe disease. Previously, we reported that loss of a key transcriptional regulator, hypoxia-inducible factor 1α (HIF-1α), in macrophages led to a lethal failure to control growth of Histoplasma (R. A. Fecher, M. C. Horwath, D. Friedrich, J. Rupp, G. S. Deepe, J Immunol 197:565-579, 2016, https://doi.org/10.4049/jimmunol.1600342). Inhibition of phagocyte activation due to excessive interleukin 10 by HIF-1α-deficient macrophages drove this outcome. In this study, we demonstrate that HIF-1α maintains contextually appropriate TCA cycle metabolism within Histoplasma-infected macrophages. The absence of HIF-1α results in excessive fumarate production that alters miRNA-27a regulation of interleukin-10. HIF-1α thus preserves the capacity of macrophages to transition from a permissive intracellular niche to the site of pathogen killing.
Keywords: Histoplasma; hypoxia inducible factor 1; innate immunity; lung; mitochondrial metabolism.
Figures



References
-
- Feng D, Sangster-Guity N, Stone R, Korczeniewska J, Mancl ME, Fitzgerald-Bocarsly P, Barnes BJ. 2010. Differential requirement of histone acetylase and deacetylase activities for IRF5-mediated proinflammatory cytokine expression. J Immunol 185:6003–6012. doi:10.4049/jimmunol.1000482. - DOI - PMC - PubMed
-
- Galván-Peña S, Carroll RG, Newman C, Hinchy EC, Palsson-McDermott E, Robinson EK, Covarrubias S, Nadin A, James AM, Haneklaus M, Carpenter S, Kelly VP, Murphy MP, Modis LK, O'Neill LA. 2019. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat Commun 10:338. doi:10.1038/s41467-018-08187-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
