Mutation in Abl kinase with altered drug-binding kinetics indicates a novel mechanism of imatinib resistance
- PMID: 34750265
- PMCID: PMC8609647
- DOI: 10.1073/pnas.2111451118
Mutation in Abl kinase with altered drug-binding kinetics indicates a novel mechanism of imatinib resistance
Abstract
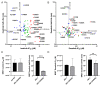
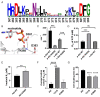
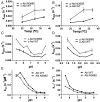
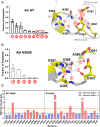
Protein kinase inhibitors are potent anticancer therapeutics. For example, the Bcr-Abl kinase inhibitor imatinib decreases mortality for chronic myeloid leukemia by 80%, but 22 to 41% of patients acquire resistance to imatinib. About 70% of relapsed patients harbor mutations in the Bcr-Abl kinase domain, where more than a hundred different mutations have been identified. Some mutations are located near the imatinib-binding site and cause resistance through altered interactions with the drug. However, many resistance mutations are located far from the drug-binding site, and it remains unclear how these mutations confer resistance. Additionally, earlier studies on small sets of patient-derived imatinib resistance mutations indicated that some of these mutant proteins were in fact sensitive to imatinib in cellular and biochemical studies. Here, we surveyed the resistance of 94 patient-derived Abl kinase domain mutations annotated as disease relevant or resistance causing using an engagement assay in live cells. We found that only two-thirds of mutations weaken imatinib affinity by more than twofold compared to Abl wild type. Surprisingly, one-third of mutations in the Abl kinase domain still remain sensitive to imatinib and bind with similar or higher affinity than wild type. Intriguingly, we identified three clinical Abl mutations that bind imatinib with wild type-like affinity but dissociate from imatinib considerably faster. Given the relevance of residence time for drug efficacy, mutations that alter binding kinetics could cause resistance in the nonequilibrium environment of the body where drug export and clearance play critical roles.
Keywords: binding kinetics; drug binding; drug resistance; imatinib; protein kinase.
Figures

References
-
- Jemal A., et al. ., Cancer statistics, 2007. CA Cancer J. Clin. 57, 43–66 (2007). - PubMed
-
- Parker S. L., Tong T., Bolden S., Wingo P. A., Cancer statistics, 1997. CA Cancer J. Clin. 47, 5–27 (1997). - PubMed
-
- Gorre M. E., et al. ., Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880 (2001). - PubMed
-
- Azam M., Latek R. R., Daley G. Q., Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell 112, 831–843 (2003). - PubMed
-
- Shah N. P., et al. ., Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2, 117–125 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

