In vitro reconstitution of the Escherichia coli 70S ribosome with a full set of recombinant ribosomal proteins
- PMID: 34750629
- PMCID: PMC8863084
- DOI: 10.1093/jb/mvab121
In vitro reconstitution of the Escherichia coli 70S ribosome with a full set of recombinant ribosomal proteins
Erratum in
-
Erratum.J Biochem. 2022 Mar 31;171(4):469. doi: 10.1093/jb/mvac016. J Biochem. 2022. PMID: 35181785 Free PMC article. No abstract available.
Abstract
Many studies of the reconstitution of the Escherichia coli small ribosomal subunit from its individual molecular parts have been reported, but contrastingly, similar studies of the large ribosomal subunit have not been well performed to date. Here, we describe protocols for preparing the 33 ribosomal proteins of the E. coli 50S subunit and demonstrate successful reconstitution of a functionally active 50S particle that can perform protein synthesis in vitro. We also successfully reconstituted both ribosomal subunits (30S and 50S) and 70S ribosomes using a full set of recombinant ribosomal proteins by integrating our developed method with the previously developed fully recombinant-based integrated synthesis, assembly and translation. The approach described here makes a major contribution to the field of ribosome engineering and could be fundamental to the future studies of ribosome assembly processes.
Keywords: PURE system; cell-free protein synthesis; protein translation; ribosomal protein; ribosome assembly.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Japanese Biochemical Society. All rights reserved.
Figures
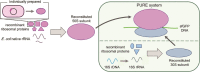





References
-
- Mizushima, S., and Nomura, M. (1970) Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226, 1214–1218 - PubMed
-
- Nikolay, R., Hilal, T., Qin, B., Mielke, T., Bürger, J., Loerke, J., Textoris-Taube, K., Nierhaus, K.H., and Spahn, C.M.T. (2018) Structural visualization of the formation and activation of the 50S ribosomal subunit during in vitro reconstitution. Mol. Cell 70, 881–893.e3 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

