This is a preprint.
Camel nanobodies broadly neutralize SARS-CoV-2 variants
- PMID: 34751270
- PMCID: PMC8575140
- DOI: 10.1101/2021.10.27.465996
Camel nanobodies broadly neutralize SARS-CoV-2 variants
Update in
-
Dromedary camel nanobodies broadly neutralize SARS-CoV-2 variants.Proc Natl Acad Sci U S A. 2022 May 3;119(18):e2201433119. doi: 10.1073/pnas.2201433119. Epub 2022 Apr 27. Proc Natl Acad Sci U S A. 2022. PMID: 35476528 Free PMC article.
Abstract
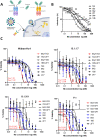
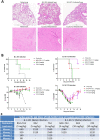
With the emergence of SARS-CoV-2 variants, there is urgent need to develop broadly neutralizing antibodies. Here, we isolate two V H H nanobodies (7A3 and 8A2) from dromedary camels by phage display, which have high affinity for the receptor-binding domain (RBD) and broad neutralization activities against SARS-CoV-2 and its emerging variants. Cryo-EM complex structures reveal that 8A2 binds the RBD in its up mode and 7A3 inhibits receptor binding by uniquely targeting a highly conserved and deeply buried site in the spike regardless of the RBD conformational state. 7A3 at a dose of ≥5 mg/kg efficiently protects K18-hACE2 transgenic mice from the lethal challenge of B.1.351 or B.1.617.2, suggesting that the nanobody has promising therapeutic potentials to curb the COVID-19 surge with emerging SARS-CoV-2 variants.
One-sentence summary: Dromedary camel ( Camelus dromedarius ) V H H phage libraries were built for isolation of the nanobodies that broadly neutralize SARS-CoV-2 variants.
Conflict of interest statement
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
