Evaluation of Efficient Non-reducing Enzymatic and Chemical Ligation Strategies for Complex Disulfide-Rich Peptides
- PMID: 34751572
- PMCID: PMC10167913
- DOI: 10.1021/acs.bioconjchem.1c00452
Evaluation of Efficient Non-reducing Enzymatic and Chemical Ligation Strategies for Complex Disulfide-Rich Peptides
Abstract
Double-knotted peptides identified in venoms and synthetic bivalent peptide constructs targeting ion channels are emerging tools for the study of ion channel pharmacology and physiology. These highly complex and disulfide-rich peptides contain two individual cystine knots, each comprising six cysteines and three disulfide bonds. Until now, native double-knotted peptides, such as Hi1a and DkTx, have only been isolated from venom or produced recombinantly, whereas engineered double-knotted peptides have successfully been produced through enzymatic ligation using sortase A to form a seamless amide bond at the ligation site between two knotted toxins, and by alkyne/azide click chemistry, joining two peptide knots via a triazole linkage. To further pursue these double-knotted peptides as pharmacological tools or probes for therapeutically relevant ion channels, we sought to identify a robust methodology resulting in a high yield product that lends itself to rapid production and facile mutational studies. In this study, we evaluated the ligation efficiency of enzymatic (sortase A5°, butelase 1, wild-type OaAEP 1, C247A-OaAEP 1, and peptiligase) and mild chemical approaches (α-ketoacid-hydroxylamine, KAHA) for forming a native amide bond linking the toxins while maintaining the native disulfide connectivity of each pre-folded peptide. We used two NaV1.7 inhibitors: PaurTx3, a spider-derived gating modifier peptide, and KIIIA, a small cone snail-derived pore blocker peptide, which have previously been shown to increase affinity and inhibitory potency on hNaV1.7 when ligated together. Correctly folded peptides were successfully ligated in varying yields, without disulfide bond shuffling or reduction, with sortase A5° being the most efficient, resulting in 60% ligation conversion within 15 min. In addition, electrophysiology studies demonstrated that for these two peptides, the amino acid composition of the linker did not affect the activity of the double-knotted peptides. This study demonstrates the powerful application of enzymes in efficiently ligating complex disulfide-rich peptides, paving the way for facile production of double-knotted peptides.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


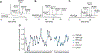
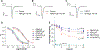
Similar articles
-
Enzymatic Ligation of Disulfide-Rich Animal Venom Peptides: Using Sortase A to Form Double-Knotted Peptides.Methods Mol Biol. 2021;2355:83-92. doi: 10.1007/978-1-0716-1617-8_8. Methods Mol Biol. 2021. PMID: 34386952 Free PMC article.
-
Enzymatic Ligation of a Pore Blocker Toxin and a Gating Modifier Toxin: Creating Double-Knotted Peptides with Improved Sodium Channel NaV1.7 Inhibition.Bioconjug Chem. 2020 Jan 15;31(1):64-73. doi: 10.1021/acs.bioconjchem.9b00744. Epub 2019 Dec 16. Bioconjug Chem. 2020. PMID: 31790574
-
Efficient Enzymatic Ligation of Inhibitor Cystine Knot Spider Venom Peptides: Using Sortase A To Form Double-Knottins That Probe Voltage-Gated Sodium Channel NaV1.7.Bioconjug Chem. 2018 Oct 17;29(10):3309-3319. doi: 10.1021/acs.bioconjchem.8b00505. Epub 2018 Sep 12. Bioconjug Chem. 2018. PMID: 30148615
-
Engineered Sortases in Peptide and Protein Chemistry.Chembiochem. 2021 Apr 16;22(8):1347-1356. doi: 10.1002/cbic.202000745. Epub 2021 Feb 3. Chembiochem. 2021. PMID: 33290621 Free PMC article. Review.
-
Sortase-mediated backbone cyclization of proteins and peptides.Biol Chem. 2015 Apr;396(4):283-93. doi: 10.1515/hsz-2014-0260. Biol Chem. 2015. PMID: 25581753 Review.
Cited by
-
Synthesis of full-length homodimer αD-VxXXB that targets human α7 nicotinic acetylcholine receptors.RSC Med Chem. 2022 Sep 5;13(11):1410-1419. doi: 10.1039/d2md00188h. eCollection 2022 Nov 16. RSC Med Chem. 2022. PMID: 36439982 Free PMC article.
References
-
- Chassagnon IR; McCarthy CA; Chin YKY; Pineda SS; Keramidas A; Mobli M; Pham V; de Silva TM; Lynch JW; Widdop RE; Rash LD; King GF Potent neuroprotection after stroke afforded by a double-knot spider-venom peptide that inhibits acid-sensing ion channel 1a. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 3750–3755. - PMC - PubMed
-
- Vassilevski AA; Fedorova IM; Maleeva EE; Korolkova YV; Efimova SS; Samsonova OV; Schagina LV; Feofanov AV; Magazanik LG; Grishin EV Novel class of spider toxin: active principle from the yellow sac spider Cheiracanthium punctorium venom is a unique two-domain polypeptide. J. Biol. Chem 2010, 285, 32293. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

