Use of tandem affinity-buffer exchange chromatography online with native mass spectrometry for optimizing overexpression and purification of recombinant proteins
- PMID: 34752295
- PMCID: PMC9055603
- DOI: 10.1016/bs.mie.2021.07.007
Use of tandem affinity-buffer exchange chromatography online with native mass spectrometry for optimizing overexpression and purification of recombinant proteins
Abstract
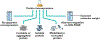
Purification of recombinant proteins typically entails overexpression in heterologous systems and subsequent chromatography-based isolation. While denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis is routinely used to screen a variety of overexpression conditions (e.g., host, medium, inducer concentration, post-induction temperature and/or incubation time) and to assess the purity of the final product, its limitations, including aberrant protein migration due to compositional eccentricities or incomplete denaturation, often preclude firm conclusions regarding the extent of overexpression and/or purification. Therefore, we recently reported an automated liquid chromatography-mass spectrometry-based strategy that couples immobilized metal affinity chromatography (IMAC) with size exclusion-based online buffer exchange (OBE) and native mass spectrometry (nMS) to directly analyze cell lysates for the presence of target proteins. IMAC-OBE-nMS can be used to assess whether target proteins (1) are overexpressed in soluble form, (2) bind and elute from an IMAC resin, (3) oligomerize, and (4) have the expected mass. Here, we use four poly-His-tagged proteins to demonstrate the potential of IMAC-OBE-nMS for expedient optimization of overexpression and purification conditions for recombinant protein production.
Keywords: Immobilized metal affinity chromatography; Liquid chromatography–mass spectrometry; Native mass spectrometry; Online buffer exchange; Protein overexpression.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest statement The desalting cartridge used in this study is a prototype of a commercial product that is being developed by Thermo Scientific with input and testing by the OSU team.
Figures
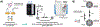
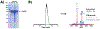
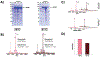
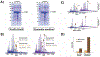
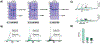
References
-
- Ali MH, & Imperiali B (2005). Protein oligomerization: how and why. Bioorg. Med. Chem, 13(17), 5013–5020. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

