Gene-corrected p.A30P SNCA patient-derived isogenic neurons rescue neuronal branching and function
- PMID: 34754035
- PMCID: PMC8578337
- DOI: 10.1038/s41598-021-01505-x
Gene-corrected p.A30P SNCA patient-derived isogenic neurons rescue neuronal branching and function
Abstract
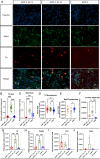
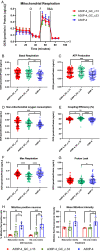
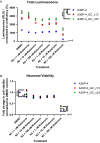
Parkinson's disease (PD) is characterised by the degeneration of A9 dopaminergic neurons and the pathological accumulation of alpha-synuclein. The p.A30P SNCA mutation generates the pathogenic form of the alpha-synuclein protein causing an autosomal-dominant form of PD. There are limited studies assessing pathogenic SNCA mutations in patient-derived isogenic cell models. Here we provide a functional assessment of dopaminergic neurons derived from a patient harbouring the p.A30P SNCA mutation. Using two clonal gene-corrected isogenic cell lines we identified image-based phenotypes showing impaired neuritic processes. The pathological neurons displayed impaired neuronal activity, reduced mitochondrial respiration, an energy deficit, vulnerability to rotenone, and transcriptional alterations in lipid metabolism. Our data describes for the first time the mutation-only effect of the p.A30P SNCA mutation on neuronal function, supporting the use of isogenic cell lines in identifying image-based pathological phenotypes that can serve as an entry point for future disease-modifying compound screenings and drug discovery strategies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

