Chikungunya virus molecular evolution in India since its re-emergence in 2005
- PMID: 34754512
- PMCID: PMC8570154
- DOI: 10.1093/ve/veab074
Chikungunya virus molecular evolution in India since its re-emergence in 2005
Abstract
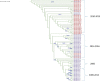
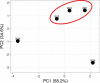
Chikungunya virus (CHIKV), an alphavirus of the Togaviridae family, is among the most medically significant mosquito-borne viruses, capable of causing major epidemics of febrile disease and severe, chronic arthritis. Identifying viral mutations is crucial for understanding virus evolution and evaluating those genetic determinants that directly impact pathogenesis and transmissibility. The present study was undertaken to expand on past CHIKV evolutionary studies through robust genome-scale phylogenetic analysis to better understand CHIKV genetic diversity and evolutionary dynamics since its reintroduction into India in 2005. We sequenced the complete genomes of fifty clinical isolates collected between 2010 and 2016 from two geographic locations, Delhi and Mumbai. We then analysed them along with 753 genomes available on the Virus Pathogen Database and Analysis Resource sampled over fifteen years (2005-20) from a range of locations across the globe and identified novel genetic variants present in samples from this study. Our analyses show evidence of frequent reintroduction of the virus into India and that the most recent CHIKV outbreak shares a common ancestor as recently as 2006.
Keywords: chikungunya virus (CHIKV); phylogeny; variants; whole-genome sequencing.
© The Author(s) 2021. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Agarwal A. et al. (2016) ‘Two Novel Epistatic Mutations (E1: K211E and E2: V264A) in Structural Proteins of Chikungunya Virus Enhance Fitness in Aedes aegypti’, Virology, 497: S59–68. - PubMed
-
- Arankalle V. A. et al. (2007) ‘Genetic Divergence of Chikungunya Viruses in India (1963–2006) with Special Reference to the 2005–2006 Explosive Epidemic’, The Journal of General Virology, 88: 1967–76. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

