Platelet transcriptome identifies progressive markers and potential therapeutic targets in chronic myeloproliferative neoplasms
- PMID: 34755136
- PMCID: PMC8561315
- DOI: 10.1016/j.xcrm.2021.100425
Platelet transcriptome identifies progressive markers and potential therapeutic targets in chronic myeloproliferative neoplasms
Abstract
Predicting disease progression remains a particularly challenging endeavor in chronic degenerative disorders and cancer, thus limiting early detection, risk stratification, and preventive interventions. Here, profiling the three chronic subtypes of myeloproliferative neoplasms (MPNs), we identify the blood platelet transcriptome as a proxy strategy for highly sensitive progression biomarkers that also enables prediction of advanced disease via machine-learning algorithms. The MPN platelet transcriptome reveals an incremental molecular reprogramming that is independent of patient driver mutation status or therapy. Subtype-specific markers offer mechanistic and therapeutic insights, and highlight impaired proteostasis and a persistent integrated stress response. Using a LASSO model with validation in two independent cohorts, we identify the advanced subtype MF at high accuracy and offer a robust progression signature toward clinical translation. Our platelet transcriptome snapshot of chronic MPNs demonstrates a proof-of-principle for disease risk stratification and progression beyond genetic data alone, with potential utility in other progressive disorders.
Keywords: MPN; biomarker; blood platelets; myeloproliferative neoplasms; platelet RNA-seq; platelet transcriptome; prediction algorithms; progression signatures; proteostasis; ruxolitinib.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
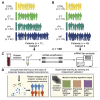


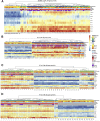
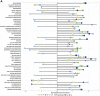

Comment in
-
Message in a platelet: decoding platelet transcriptomes in myeloproliferative neoplasms.Cell Rep Med. 2021 Oct 19;2(10):100433. doi: 10.1016/j.xcrm.2021.100433. eCollection 2021 Oct 19. Cell Rep Med. 2021. PMID: 34755142 Free PMC article.
References
-
- Rondina M.T., Voora D., Simon L.M., Schwertz H., Harper J.F., Lee O., Bhatlekar S.C., Li Q., Eustes A.S., Montenont E. Longitudinal RNA-seq analysis of the repeatability of gene expression and splicing in human platelets identifies a platelet SELP splice QTL. Circ. Res. 2020;126:501–516. - PMC - PubMed
-
- Finazzi G., De Stefano V., Barbui T. Are MPNs vascular diseases? Curr. Hematol. Malig. Rep. 2013;8:307–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

