Rhea, the reaction knowledgebase in 2022
- PMID: 34755880
- PMCID: PMC8728268
- DOI: 10.1093/nar/gkab1016
Rhea, the reaction knowledgebase in 2022
Abstract
Rhea (https://www.rhea-db.org) is an expert-curated knowledgebase of biochemical reactions based on the chemical ontology ChEBI (Chemical Entities of Biological Interest) (https://www.ebi.ac.uk/chebi). In this paper, we describe a number of key developments in Rhea since our last report in the database issue of Nucleic Acids Research in 2019. These include improved reaction coverage in Rhea, the adoption of Rhea as the reference vocabulary for enzyme annotation in the UniProt knowledgebase UniProtKB (https://www.uniprot.org), the development of a new Rhea website, and the designation of Rhea as an ELIXIR Core Data Resource. We hope that these and other developments will enhance the utility of Rhea as a reference resource to study and engineer enzymes and the metabolic systems in which they function.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

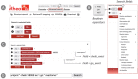

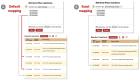
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

