A Novel Humanized Model of NASH and Its Treatment With META4, A Potent Agonist of MET
- PMID: 34756982
- PMCID: PMC8688725
- DOI: 10.1016/j.jcmgh.2021.10.007
A Novel Humanized Model of NASH and Its Treatment With META4, A Potent Agonist of MET
Abstract
Background & aims: Nonalcoholic fatty liver disease is a frequent cause of hepatic dysfunction and is now a global epidemic. This ailment can progress to an advanced form called nonalcoholic steatohepatitis (NASH) and end-stage liver disease. Currently, the molecular basis of NASH pathogenesis is poorly understood, and no effective therapies exist to treat NASH. These shortcomings are due to the paucity of experimental NASH models directly relevant to humans.
Methods: We used chimeric mice with humanized liver to investigate nonalcoholic fatty liver disease in a relevant model. We carried out histologic, biochemical, and molecular approaches including RNA-Seq. For comparison, we used side-by-side human NASH samples.
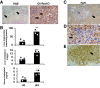
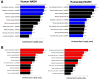
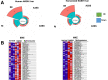
Results: Herein, we describe a "humanized" model of NASH using transplantation of human hepatocytes into fumarylacetoacetate hydrolase-deficient mice. Once fed a high-fat diet, these mice develop NAFLD faithfully, recapitulating human NASH at the histologic, cellular, biochemical, and molecular levels. Our RNA-Seq analyses uncovered that a variety of important signaling pathways that govern liver homeostasis are profoundly deregulated in both humanized and human NASH livers. Notably, we made the novel discovery that hepatocyte growth factor (HGF) function is compromised in human and humanized NASH at several levels including a significant increase in the expression of the HGF antagonists known as NK1/NK2 and marked decrease in HGF activator. Based on these observations, we generated a potent, human-specific, and stable agonist of human MET that we have named META4 (Metaphor) and used it in the humanized NASH model to restore HGF function.
Conclusions: Our studies revealed that the humanized NASH model recapitulates human NASH and uncovered that HGF-MET function is impaired in this disease. We show that restoring HGF-MET function by META4 therapy ameliorates NASH and reinstates normal liver function in the humanized NASH model. Our results show that the HGF-MET signaling pathway is a dominant regulator of hepatic homeostasis.
Keywords: FAH Mice; Fatty Liver Disease; HGF; HGF antagonist; Hepatocyte Growth Factor; High-fat Diet; Humanized Liver; Liver Cancer; MET; Metabolic Syndrome; NAFLD; NASH; NK1; NK2; Type 2 Diabetes.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures















References
-
- Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease: meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. - PubMed
-
- Rinella M., Charlton M. The globalization of nonalcoholic fatty liver disease: prevalence and impact on world health. Hepatology. 2016;64:19–22. - PubMed
-
- Anstee Q.M., Targher G., Day C.P. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330–344. - PubMed
-
- Smith B.W., Adams L.A. Nonalcoholic fatty liver disease and diabetes mellitus: pathogenesis and treatment. Nat Rev Endocrinol. 2011;7:456–465. - PubMed
-
- Hardy T., Oakley F., Anstee Q.M., Day C.P. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu Rev Pathol. 2016;11:451–496. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

